committed by
 GitHub
GitHub
No known key found for this signature in database
GPG Key ID: 4AEE18F83AFDEB23
85 changed files with 1530877 additions and 3 deletions
Unified View
Diff Options
-
+23 -0ML Cookbook/BasicNeuralNet.py
-
+16 -0ML Cookbook/CNNs/BasicCNN.py
-
+342 -0ML Cookbook/DecisionTree.py
-
+29 -0ML Cookbook/clean_text.py
-
+41 -0ML Cookbook/perceptron.py
-
+41 -0ML Cookbook/perceptron1.py
-
+41 -0ML Cookbook/text_preprocessing
-
+63 -0bfs.py
-
+120 -0config/vimrc
-
+58 -0dfs.py
-
+579 -0geneticAlgorithm/geneticAlgo.html
-
+92 -0graphing/basicGraph.html
-
+1 -0graphing/ex.json
-
+142 -0graphing/floyd_warshall.py
-
+122 -0graphing/graph.py
-
+71 -0multiThreadedFileIO/DataCreator.java
-
+44 -0multiThreadedFileIO/FileReader.java
-
+40 -0multiThreadedFileIO/MultiThreadedFileReadTest.java
-
+20 -0multiThreadedFileIO/ReadTask.java
-
+87 -0multiThreadedFileIO/TaskManager.java
-
+13 -0multiThreadedFileIO/graph.py
-
+306 -0notebooks/8-bit/8-bit.ipynb
-
BINnotebooks/8-bit/dolphin.jpg
-
+507 -0notebooks/burnout/burnout.ipynb
-
+407 -0notebooks/cuda-vs-cpu.ipynb
-
+1168 -0notebooks/fitbit/GraphingMyLifeWithFitbit.ipynb
-
BINnotebooks/fitbit/compression.png
-
+129601 -0notebooks/fitbit/data/calories/calories-2019-07-01.json
-
+7486 -0notebooks/fitbit/data/sleep-2019-04-02.json
-
+8059 -0notebooks/fitbit/data/sleep-2019-06-01.json
-
+8492 -0notebooks/fitbit/data/sleep-2019-07-01.json
-
+6336 -0notebooks/fitbit/data/sleep-2019-07-31.json
-
+8372 -0notebooks/fitbit/data/sleep-2019-09-29.json
-
+8540 -0notebooks/fitbit/data/sleep-2019-11-28.json
-
+8896 -0notebooks/fitbit/data/sleep-2019-12-28.json
-
+182 -0notebooks/fitbit/data/sleep/sleep_score.csv
-
+118294 -0notebooks/fitbit/data/steps-2019-04-02.json
-
+124426 -0notebooks/fitbit/data/steps-2019-05-02.json
-
+122410 -0notebooks/fitbit/data/steps-2019-06-01.json
-
+123349 -0notebooks/fitbit/data/steps-2019-07-01.json
-
+93460 -0notebooks/fitbit/data/steps-2019-07-31.json
-
+126109 -0notebooks/fitbit/data/steps-2019-08-30.json
-
+125791 -0notebooks/fitbit/data/steps-2019-09-29.json
-
+123265 -0notebooks/fitbit/data/steps-2019-10-29.json
-
+121957 -0notebooks/fitbit/data/steps-2019-11-28.json
-
+124729 -0notebooks/fitbit/data/steps-2019-12-28.json
-
+123559 -0notebooks/fitbit/data/steps-2020-01-27.json
-
+5272 -0notebooks/fitbit/data/steps-2020-02-26.json
-
BINnotebooks/fitbit/dataExport.png
-
+428 -0notebooks/fitbit/fitbitsleep.ipynb
-
BINnotebooks/fitbit/restingHeartRate.jpg
-
+1124 -0notebooks/fitbit/restingHeartRate.svg
-
BINnotebooks/k-means/bg.jpg
-
BINnotebooks/k-means/filters.png
-
+587 -0notebooks/k-means/k-means.ipynb
-
+43 -0notebooks/node_2_vec/friendsMap.map
-
+320 -0notebooks/node_2_vec/jrtechs.edgelist
-
+39 -0notebooks/node_2_vec/output/jrtechs2.emd
-
+278 -0notebooks/node_2_vec/steam_node_2_vec.ipynb
-
+288 -0notebooks/parallel-java.ipynb
-
+1063 -0notebooks/pytorch-headfirst.ipynb
-
+1 -0notebooks/steam/games.json
-
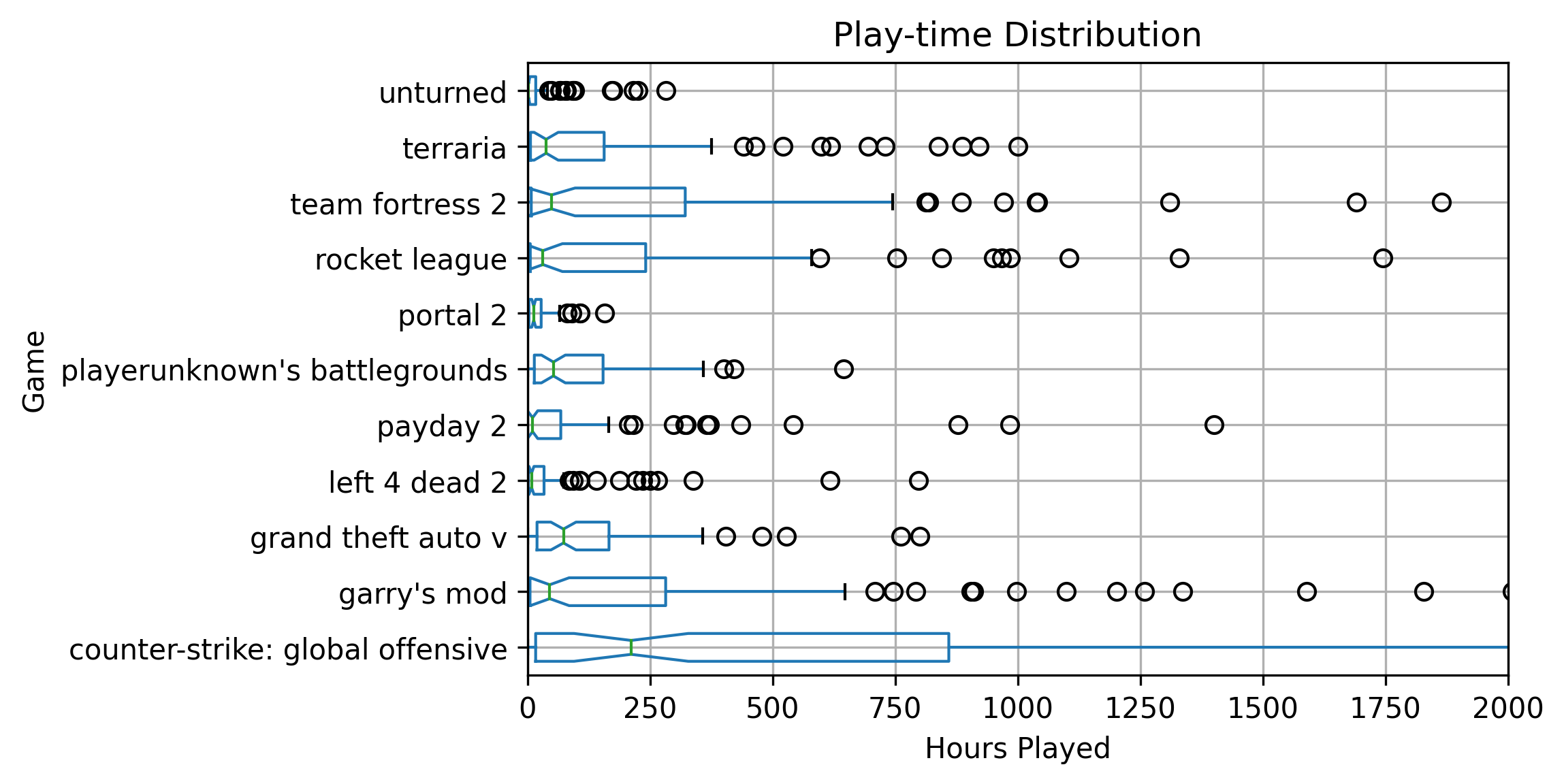
BINnotebooks/steam/playTimes.png
-
+1454 -0notebooks/steam/steam.ipynb
-
+486 -0notebooks/word-embeddings-part2/embeddings.ipynb
-
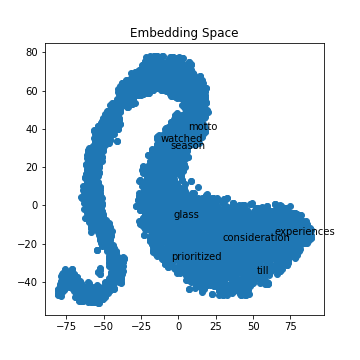
BINnotebooks/word-embeddings-part2/ex1.png
-
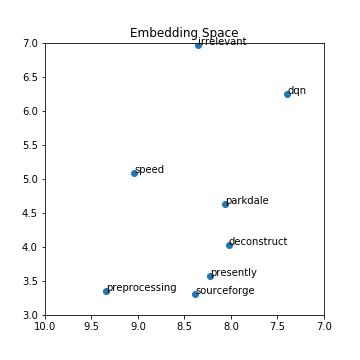
BINnotebooks/word-embeddings-part2/smallex.png
-
+284 -0notebooks/word-embeddings/Untitled.ipynb
-
+678 -0notebooks/word-embeddings/embedding-part-1.ipynb
-
BINnotebooks/word-embeddings/swap.png
-
+82 -0other/Dijkstra.py
-
+52 -0other/knapSack.py
-
+129 -0other/markdownParagraphFormatter.py
-
+14 -3other/minMul.py
-
+162 -0other/paragraphFormatting.py
-
+65 -0perceptron/perceptron.py
-
+42 -0searching_algo/BFS.py
-
+39 -0searching_algo/Jump_Search.py
-
+33 -0searching_algo/exponentialSearch.py
-
+35 -0sorting/bucketSort.py
-
+122 -0sorting/heapsort.py
-
+39 -0text_preprocessing.py
-
+14 -0windows/Sticky Notes Backuper/Readme.txt
-
+9 -0windows/Sticky Notes Backuper/grabStickyNotes.bat
-
+9 -0windows/Sticky Notes Backuper/replaceCurrentStickyNotes.bat
+ 23
- 0
ML Cookbook/BasicNeuralNet.py
View File
| @ -0,0 +1,23 @@ | |||||
| from torch import nn | |||||
| class BasicNeuralNet(nn.Module): | |||||
| def __init__(self): | |||||
| super().__init__() | |||||
| # Inputs to hidden layer linear transformation | |||||
| self.hidden = nn.Linear(784, 256) | |||||
| # Output layer, 10 units | |||||
| self.output = nn.Linear(256, 10) | |||||
| # Define sigmoid activation and softmax output | |||||
| self.sigmoid = nn.Sigmoid() | |||||
| self.softmax = nn.Softmax(dim=1) | |||||
| def forward(self, x): | |||||
| # Pass the input tensor through each of the operations | |||||
| x = self.hidden(x) | |||||
| x = self.sigmoid(x) | |||||
| x = self.output(x) | |||||
| x = self.softmax(x) | |||||
| return x | |||||
+ 16
- 0
ML Cookbook/CNNs/BasicCNN.py
View File
| @ -0,0 +1,16 @@ | |||||
| import torch.nn as nn | |||||
| import torch.nn.functional as F | |||||
| # CNN architecture definition | |||||
| class Net(nn.Module): | |||||
| def __init__(self): | |||||
| super(Net, self).__init__() | |||||
| # convolutional layer | |||||
| self.conv1 = nn.Conv2d(3, 16, 3, padding=1) | |||||
| # max pooling layer | |||||
| self.pool = nn.MaxPool2d(2, 2) | |||||
| def forward(self, x): | |||||
| # add sequence of convolutional and max pooling layers | |||||
| x = self.pool(F.relu(self.conv1(x))) | |||||
| return x | |||||
+ 342
- 0
ML Cookbook/DecisionTree.py
View File
| @ -0,0 +1,342 @@ | |||||
| """ | |||||
| :Author: James Sherratt | |||||
| :Date: 21/10/2019 | |||||
| :License: MIT | |||||
| :name: DecisionTree.py | |||||
| Basic implementation of a binary decision tree algorithm, with one | |||||
| discriminant per node. | |||||
| Useful links: | |||||
| https://scikit-learn.org/stable/modules/tree.html | |||||
| https://en.wikipedia.org/wiki/Decision_tree | |||||
| """ | |||||
| import numpy as np | |||||
| from sklearn import datasets | |||||
| def proportion_k(ym): | |||||
| """ | |||||
| Get the proportions of each class in the current set of values. | |||||
| :param ym: y values (class) of the data at a given node. | |||||
| :return: list containing the classes and the fraction of those classes present. | |||||
| """ | |||||
| counts = list(np.unique(ym, return_counts=True)) | |||||
| counts[1] = counts[1]/(ym.shape[0]) | |||||
| return counts | |||||
| def gini(k_proportions): | |||||
| """ | |||||
| Gini impurity function. This is used to determine the impurity of a given | |||||
| set of data, given the proportions of the classes in the dataset. | |||||
| This is equivalent to: | |||||
| H = ∑ pk(1-pk) for all k classes. | |||||
| k_proportions, in this case, is an array of pk's | |||||
| :param k_proportions: array containing proportions of different classes. Proportions sum to 1. | |||||
| :return: the impurity of the dataset. | |||||
| """ | |||||
| return (k_proportions*(1-k_proportions)).sum() | |||||
| def node_impurity(ym): | |||||
| """ | |||||
| Calculate the impurity of data on one side of node after split. | |||||
| :param ym: Actual y data for the selected dataset. | |||||
| :return: dict containing the impurity value of the side and the most common class on that side. | |||||
| """ | |||||
| if ym.shape[0] == 0: | |||||
| return {"impurity": 0, "max_class": 0} | |||||
| k_prop = proportion_k(ym) | |||||
| return {"impurity": gini(k_prop[1]), "max_class": k_prop[0][np.argmax(k_prop[1])]} | |||||
| def disc_val_impurity(yleft, yright): | |||||
| """ | |||||
| Calculate the level of impurity left in the given data after splitting. This returns | |||||
| a dict which contains: | |||||
| - The impurity of the data after being split. | |||||
| - The class of the largest proportion on the left and right side of the split. | |||||
| The aim is to find a split which minimises impurity. | |||||
| The impurity calculated is: | |||||
| G = (nleft/ntot)*Hleft + (nright/ntot)*Hright | |||||
| This gives the impurity of the split data. | |||||
| :param yleft: Real/ training y values for the data on the left. | |||||
| :param yright: Real/ training y values for the data on the right. | |||||
| :return: Dict containing the data impurity after split and the most common class on the left and right of the split. | |||||
| """ | |||||
| nleft = yleft.shape[0] | |||||
| nright = yright.shape[0] | |||||
| ntot = nleft + nright | |||||
| left_imp = node_impurity(yleft) | |||||
| right_imp = node_impurity(yright) | |||||
| return { | |||||
| "impurity": ((nleft/ntot)*left_imp["impurity"])+((nright/ntot)*right_imp["impurity"]), | |||||
| "lmax_class": left_imp["max_class"], | |||||
| "rmax_class": right_imp["max_class"] | |||||
| } | |||||
| def niave_min_impurity(xm, ym): | |||||
| """ | |||||
| Find a discriminator which minimises the impurity of the data. The discriminator | |||||
| is used to split data at a node. | |||||
| This works by: | |||||
| 1. Selecting a data column as a discriminator. | |||||
| 2. Splitting the possible values of the discriminator into 1000 even spaced values | |||||
| (between the minimum and maximum value in the dataset). | |||||
| 3. Selecting the discriminator column + value which minimises the impurity. | |||||
| :param xm: Data on the left. | |||||
| :param ym: Data on the right. | |||||
| :return: dict containing the current niave minimum impurity. | |||||
| """ | |||||
| minxs = xm.min(axis=0) | |||||
| maxxs = xm.max(axis=0) | |||||
| # discriminator with the smallest impurity. | |||||
| cur_min_disc = None | |||||
| # Choose a column to discriminate by. | |||||
| for x_idx, (dmin, dmax) in enumerate(zip(minxs, maxxs)): | |||||
| # Create a set of possibly values to use as the discriminator for that column. | |||||
| disc_vals = np.linspace(dmin, dmax, 1000) | |||||
| for disc_val in disc_vals: | |||||
| selection = xm[:, x_idx] < disc_val | |||||
| yleft = ym[selection] | |||||
| yright = ym[selection==False] | |||||
| # Calculate impurity. | |||||
| imp = disc_val_impurity(yleft, yright) | |||||
| # Choose a column with the smallest impurity. | |||||
| try: | |||||
| if cur_min_disc["impurity"] > imp["impurity"]: | |||||
| imp["discriminator"] = x_idx | |||||
| imp["val"] = disc_val | |||||
| cur_min_disc = imp | |||||
| except TypeError: | |||||
| imp["discriminator"] = x_idx | |||||
| imp["val"] = disc_val | |||||
| cur_min_disc = imp | |||||
| return cur_min_disc | |||||
| class BinaryTreeClassifier: | |||||
| def __init__(self, max_depth=4, min_data=5): | |||||
| """ | |||||
| Initialise the binary decision tree classifier. This classifier works by: | |||||
| - Splitting the data into 2 sets at every node. | |||||
| - These 2 sets are then split into 2 more sets at their nodes etc. until they reach a leaf. | |||||
| - At the leaves, the data is classified into whatever class was "most common" in that leaf during training. | |||||
| :param max_depth: The maximum depth the binary tree classifier goes to. | |||||
| :param min_data: The minimum sample size of the training data before the tree stops splitting. | |||||
| """ | |||||
| tree = dict() | |||||
| self.depth = max_depth | |||||
| self.min_data = min_data | |||||
| def _node_mask(X, node): | |||||
| """ | |||||
| Get the discriminator mask for the node. This splits the data into left and right components. | |||||
| :param X: dataset input data. | |||||
| :param node: the current node of the tree, with its discriminator value. | |||||
| :return: truth array, which splits data left and right. | |||||
| """ | |||||
| return X[:, node["discriminator"]] < node["val"] | |||||
| def _apply_disc(X, y, node): | |||||
| """ | |||||
| Apply the discriminator to the data at a given node. | |||||
| :param X: dataset input. | |||||
| :param y: dataset (observed) output. | |||||
| :param node: The node to split data by. | |||||
| :return: The x and y data, split left and right. | |||||
| """ | |||||
| left_cond = BinaryTreeClassifier._node_mask(X, node) | |||||
| right_cond = left_cond == False | |||||
| left_X, left_y = X[left_cond], y[left_cond] | |||||
| right_X, right_y = X[right_cond], y[right_cond] | |||||
| return left_X, left_y, right_X, right_y | |||||
| def _tree_node(X, y, max_depth, min_data): | |||||
| """ | |||||
| Create a tree node. This also creates child nodes of this node recursively. | |||||
| :param X: input data for the dataset at a node. | |||||
| :param y: output (observed) data for the dataset at a node. | |||||
| :param max_depth: The maximum depth of the tree from this node. | |||||
| :param min_data: The minimum amount of data which can be discriminated. | |||||
| :return: The node + its children, as a dict. | |||||
| """ | |||||
| # Get the new node, as a dict. | |||||
| node = niave_min_impurity(X, y) | |||||
| # Split the data using the discriminator. | |||||
| left_X, left_y, right_X, right_y = BinaryTreeClassifier._apply_disc(X, y, node) | |||||
| if max_depth > 1: | |||||
| if left_X.shape[0] >= min_data: | |||||
| # Create a new node on the left (recursively) if max depth | |||||
| # and min data have not been reached. | |||||
| node["left"] = BinaryTreeClassifier._tree_node(left_X, left_y, max_depth-1, min_data) | |||||
| if right_X.shape[0] >= min_data: | |||||
| # Create a new node on the right (recursively) if max depth | |||||
| # and min data have not been reached. | |||||
| node["right"] = BinaryTreeClassifier._tree_node(right_X, right_y, max_depth-1, min_data) | |||||
| return node | |||||
| def _run_tree(X, node): | |||||
| """ | |||||
| Run a node of the classifier, recurisively. | |||||
| :param node: The node to run on the data. | |||||
| :return: The classified y (expected) data. | |||||
| """ | |||||
| # Setup y array. | |||||
| y = np.zeros(X.shape[0]) | |||||
| # Get the discriminator left conditional. | |||||
| left_cond = BinaryTreeClassifier._node_mask(X, node) | |||||
| # Right conditional | |||||
| right_cond = left_cond == False | |||||
| try: | |||||
| # Try to split the data further on the left side. | |||||
| y[left_cond] = BinaryTreeClassifier._run_tree(X[left_cond], node["left"]) | |||||
| except KeyError: | |||||
| # If we cannot split any further, get the class of the data on the left (as this is a leaf). | |||||
| y[left_cond] = node["lmax_class"] | |||||
| try: | |||||
| # Try to split the data further on the right side. | |||||
| y[right_cond] = BinaryTreeClassifier._run_tree(X[right_cond], node["right"]) | |||||
| except KeyError: | |||||
| # If we cannot split any further, get the class of the data on the right (as this is a leaf). | |||||
| y[right_cond] = node["rmax_class"] | |||||
| return y | |||||
| def _node_dict(node, idx=0): | |||||
| """ | |||||
| Get a dict of all the nodes, recursively. The keys are the index of an array, | |||||
| as if the array is a heap. | |||||
| :param node: The current node to add to the dict and to get children of recursively. | |||||
| :param idx: current index (key) of the node. | |||||
| :return: dict containing all the nodes retrieved. | |||||
| """ | |||||
| # Current nodes. | |||||
| nodes = {} | |||||
| node_data = {"lmax_class": node["lmax_class"], | |||||
| "rmax_class": node["rmax_class"], | |||||
| "discriminator": node["discriminator"], | |||||
| "val": node["val"]} | |||||
| nodes[idx] = node_data | |||||
| # Try to get the left nodes. | |||||
| try: | |||||
| left_idx = 2 * idx + 1 | |||||
| nodes.update(BinaryTreeClassifier._node_dict(node["left"], left_idx)) | |||||
| except KeyError: | |||||
| pass | |||||
| # Try to get the right nodes. | |||||
| try: | |||||
| right_idx = 2 * idx + 2 | |||||
| nodes.update(BinaryTreeClassifier._node_dict(node["right"], right_idx)) | |||||
| except KeyError: | |||||
| pass | |||||
| # return the dict of nodes retrieved. | |||||
| return nodes | |||||
| def build_tree(self, X, y): | |||||
| """ | |||||
| Build (train) the decision tree classifier. | |||||
| :param X: input training data. | |||||
| :param y: output training (observed) data. | |||||
| :return: None | |||||
| """ | |||||
| self.tree = BinaryTreeClassifier._tree_node(X, y, self.depth, self.min_data) | |||||
| def classify(self, X): | |||||
| """ | |||||
| Classify some data using the tree. | |||||
| :param X: Input data. | |||||
| :return: output (expected) classes of the data, or y values, for the given input. | |||||
| """ | |||||
| return BinaryTreeClassifier._run_tree(X, self.tree) | |||||
| def tree_to_heap_array(self): | |||||
| """ | |||||
| Convert the tree to a binary heap, stored in an array with standard indexing. | |||||
| i.e. a node at index i has children at 2i*1 and 2i+2 and a parent at (i-1)//2. | |||||
| :return: list containing the tree nodes. | |||||
| """ | |||||
| tree_dict = BinaryTreeClassifier._node_dict(self.tree) | |||||
| return [tree_dict[key] for key in sorted(tree_dict.keys())] | |||||
| def shuffle_split(x, y, frac=0.6): | |||||
| """ | |||||
| Shuffle and split X and y data. "frac" is the ratio of the split. | |||||
| e.g. 0.6 means 60% of the data goes into the left fraction, 40% into the right. | |||||
| Note X and y are shuffled the same, so row i in X data is still matched with row i in y after shuffle. | |||||
| :param x: X values of the data (predictor). | |||||
| :param y: y values of the data (observation). | |||||
| :param frac: fraction to split data by. | |||||
| :return: x1, y1, x2, y2 data where x1, y1 is the left fraction and x2, y2 is the right. | |||||
| """ | |||||
| data_idx = np.arange(x.shape[0]) | |||||
| sample1 = data_idx < (data_idx.max()*frac) | |||||
| np.random.shuffle(data_idx) | |||||
| np.random.shuffle(sample1) | |||||
| sample2 = sample1 == False | |||||
| x1, y1 = x[data_idx[sample1]], y[data_idx[sample1]] | |||||
| x2, y2 = x[data_idx[sample2]], y[data_idx[sample2]] | |||||
| return x1, y1, x2, y2 | |||||
| if __name__ == "__main__": | |||||
| # Set the seed for expected test results. | |||||
| np.random.seed(10) | |||||
| # Test decision tree with iris data. | |||||
| iris_data = datasets.load_iris() | |||||
| X = iris_data["data"] | |||||
| y = iris_data["target"] | |||||
| # Split iris data into test and train. | |||||
| X_train, y_train, X_test, y_test = shuffle_split(X, y) | |||||
| # create the decision tree classifier. | |||||
| classifier = BinaryTreeClassifier() | |||||
| # Train the classifier. | |||||
| classifier.build_tree(X_train, y_train) | |||||
| # Get the result when the classifier is applied to to the test data. | |||||
| result = classifier.classify(X_test) | |||||
| # Get the accuracy of the classifier. | |||||
| # accuracy = (number of correct results)/(total number of results) | |||||
| print("accuracy:", (result == y_test).sum()/(result.shape[0])) | |||||
| # convert the tree into a heap array. | |||||
| tree_arr = classifier.tree_to_heap_array() | |||||
| print("heap:") | |||||
| for i, node in enumerate(tree_arr): | |||||
| print(i, node) | |||||
+ 29
- 0
ML Cookbook/clean_text.py
View File
| @ -0,0 +1,29 @@ | |||||
| import re as clear | |||||
| class process(): | |||||
| def master_clean_text(text): | |||||
| #clean up all the html tags | |||||
| text = clear.sub(r'<.*?>','',text) | |||||
| #remove the unwanted punctation chars | |||||
| text = clear.sub(r"\\","",text) | |||||
| text = clear.sub(r"\'","",text) | |||||
| text = clear.sub(r"\"","",text) | |||||
| # coversion to lowercase to remove complexity | |||||
| text = text.strip().lower() | |||||
| #removing unwanted expressions | |||||
| unwanted = '!"\'#$%&()*+,-./:;<=>?@[\\]^_`{|}~\t\n' | |||||
| convert = dict((c," ") for c in unwanted) | |||||
| # str.maketrans() --->> creates a one to one mapping of a character to its translation/replacement. | |||||
| mapping_trans = str.maketrans(convert) | |||||
| text = text.translate(mapping_trans) | |||||
| return text | |||||
| #master_clean_text("<a> Say youre scrapping a text from you'r website !! WEll it might be swap CASE or unevened you wanna remove all the punctation's into separate WOrd !!!!</a>").split() | |||||
+ 41
- 0
ML Cookbook/perceptron.py
View File
| @ -0,0 +1,41 @@ | |||||
| import numpy as np | |||||
| class Perceptron(object): | |||||
| """Implements a perceptron network""" | |||||
| def __init__(self, input_size, lr=1, epochs=100): | |||||
| self.W = np.zeros(input_size+1) | |||||
| # add one for bias | |||||
| self.epochs = epochs | |||||
| self.lr = lr | |||||
| #activation function | |||||
| def activation_fn(self, x): | |||||
| #return (x >= 0).astype(np.float32) | |||||
| return 1 if x >= 0 else 0 | |||||
| #we need a prediction function to run an input through the perceptron and return an output. | |||||
| def predict(self, x): | |||||
| z = self.W.T.dot(x) | |||||
| a = self.activation_fn(z) | |||||
| return a | |||||
| def fit(self, X, d): | |||||
| for _ in range(self.epochs): | |||||
| for i in range(d.shape[0]): | |||||
| x = np.insert(X[i], 0, 1) | |||||
| y = self.predict(x) | |||||
| e = d[i] - y | |||||
| self.W = self.W + self.lr * e * x | |||||
| #the easiset set of data that we can provide is the AND gate. Given is set of inputs and outputs. | |||||
| if __name__ == '__main__': | |||||
| X = np.array([ | |||||
| [0, 0], | |||||
| [0, 1], | |||||
| [1, 0], | |||||
| [1, 1] | |||||
| ]) | |||||
| d = np.array([0, 0, 0, 1]) | |||||
| perceptron = Perceptron(input_size=2) | |||||
| perceptron.fit(X, d) | |||||
| print(perceptron.W) | |||||
+ 41
- 0
ML Cookbook/perceptron1.py
View File
| @ -0,0 +1,41 @@ | |||||
| import numpy as np | |||||
| class Perceptron(object): | |||||
| """Implements a perceptron network""" | |||||
| def __init__(self, input_size, lr=1, epochs=100): | |||||
| self.W = np.zeros(input_size+1) | |||||
| # add one for bias | |||||
| self.epochs = epochs | |||||
| self.lr = lr | |||||
| #activation function | |||||
| def activation_fn(self, x): | |||||
| #return (x >= 0).astype(np.float32) | |||||
| return 1 if x >= 0 else 0 | |||||
| #we need a prediction function to run an input through the perceptron and return an output. | |||||
| def predict(self, x): | |||||
| z = self.W.T.dot(x) | |||||
| a = self.activation_fn(z) | |||||
| return a | |||||
| def fit(self, X, d): | |||||
| for _ in range(self.epochs): | |||||
| for i in range(d.shape[0]): | |||||
| x = np.insert(X[i], 0, 1) | |||||
| y = self.predict(x) | |||||
| e = d[i] - y | |||||
| self.W = self.W + self.lr * e * x | |||||
| #the easiset set of data that we can provide is the AND gate. Given is set of inputs and outputs. | |||||
| if __name__ == '__main__': | |||||
| X = np.array([ | |||||
| [0, 0], | |||||
| [0, 1], | |||||
| [1, 0], | |||||
| [1, 1] | |||||
| ]) | |||||
| d = np.array([0, 0, 0, 1]) | |||||
| perceptron = Perceptron(input_size=2) | |||||
| perceptron.fit(X, d) | |||||
| print(perceptron.W) | |||||
+ 41
- 0
ML Cookbook/text_preprocessing
View File
| @ -0,0 +1,41 @@ | |||||
| import shutil | |||||
| from shutil import copyfile | |||||
| copyfile(src = "../input/cleantext/cleantext.py", dst = "../working/cleantext.py") | |||||
| # import all our functions | |||||
| from cleantext import * | |||||
| #!pylint cleantext | |||||
| import pandas as pd | |||||
| from sklearn.feature_extraction.text import CountVectorizer | |||||
| training = [ | |||||
| " I am master of all", | |||||
| "I am a absolute learner" | |||||
| ] | |||||
| generalization = [ | |||||
| "I am absolute learner learner" | |||||
| ] | |||||
| vectorization = CountVectorizer( | |||||
| stop_words = "english", | |||||
| preprocessor = process.master_clean_text) | |||||
| vectorization.fit(training) | |||||
| build_vocab = { | |||||
| value:key | |||||
| for key , value in vectorization.vocabulary_.items() | |||||
| } | |||||
| vocab = [build_vocab[i] for i in range(len(build_vocab))] | |||||
| pd.DataFrame( | |||||
| data = vectorization.transform(generalization).toarray(), | |||||
| index=["generalization"], | |||||
| columns=vocab | |||||
| ) | |||||
+ 63
- 0
bfs.py
View File
| @ -0,0 +1,63 @@ | |||||
| # Python3 Program to print BFS traversal | |||||
| from collections import defaultdict | |||||
| # This class represents a directed graph | |||||
| # using adjacency list representation | |||||
| class Graph: | |||||
| # Constructor | |||||
| def __init__(self): | |||||
| # default dictionary to store graph | |||||
| self.graph = defaultdict(list) | |||||
| # function to add an edge to graph | |||||
| def addEdge(self,u,v): | |||||
| self.graph[u].append(v) | |||||
| # Function to print a BFS of graph | |||||
| def BFS(self, s): | |||||
| # Mark all the vertices as not visited | |||||
| visited = [False] * (len(self.graph)) | |||||
| # Create a queue for BFS | |||||
| queue = [] | |||||
| # Mark the source node as | |||||
| # visited and enqueue it | |||||
| queue.append(s) | |||||
| visited[s] = True | |||||
| while queue: | |||||
| # Dequeue a vertex from | |||||
| # queue and print it | |||||
| s = queue.pop(0) | |||||
| print (s, end = " ") | |||||
| # Get all adjacent vertices of the | |||||
| # dequeued vertex s. If a adjacent | |||||
| # has not been visited, then mark it | |||||
| # visited and enqueue it | |||||
| for i in self.graph[s]: | |||||
| if visited[i] == False: | |||||
| queue.append(i) | |||||
| visited[i] = True | |||||
| # Driver code | |||||
| g = Graph() | |||||
| g.addEdge(0, 1) | |||||
| g.addEdge(0, 2) | |||||
| g.addEdge(1, 2) | |||||
| g.addEdge(2, 0) | |||||
| g.addEdge(2, 3) | |||||
| g.addEdge(3, 3) | |||||
| print ("Following is Breadth First Traversal" | |||||
| " (starting from vertex 2)") | |||||
| g.BFS(2) | |||||
+ 120
- 0
config/vimrc
View File
| @ -0,0 +1,120 @@ | |||||
| " sets spell check to be enabled to files which | |||||
| " end with either .md or .txt | |||||
| " | |||||
| " To get auto complete type z= when you are | |||||
| " over the word. | |||||
| autocmd BufRead,BufNewFile *.md setlocal spell spelllang=en_us | |||||
| autocmd BufRead,BufNewFile *.txt setlocal spell spelllang=en_us | |||||
| """ Indentation and Tabs """ | |||||
| "file based indentation | |||||
| filetype plugin indent on | |||||
| "copy indentation from current line when making a new line | |||||
| set autoindent | |||||
| " Smart indentation when programming: indent after { | |||||
| set smartindent | |||||
| set tabstop=4 " number of spaces per tab | |||||
| set expandtab " convert tabs to spaces | |||||
| set shiftwidth=4 " set a tab press equal to 4 spaces | |||||
| """ Looks and Appearance""" | |||||
| " Enable syntax highlighting | |||||
| syntax enable | |||||
| " Enable 256 colors palette in Gnome Terminal | |||||
| if $COLORTERM == 'gnome-terminal' | |||||
| set t_Co=256 | |||||
| endif | |||||
| try | |||||
| colorscheme desert | |||||
| catch | |||||
| endtry | |||||
| set background=dark | |||||
| " Set extra options when running in GUI mode | |||||
| if has("gui_running") | |||||
| set guioptions-=T | |||||
| set guioptions-=e | |||||
| set t_Co=256 | |||||
| set guitablabel=%M\ %t | |||||
| endif | |||||
| " File Encodings | |||||
| " Set utf8 as standard encoding and en_US as the standard language | |||||
| set encoding=utf8 | |||||
| " Use Unix as the standard file type | |||||
| set ffs=unix,dos,mac | |||||
| " Productivity | |||||
| " Set Line Numbers to show | |||||
| set number | |||||
| " Highlights the current line with a underscore | |||||
| set cursorline | |||||
| " Displays a red bar at 80 characters | |||||
| set colorcolumn=80 | |||||
| " Shows a auto complete tab when you are typing a command | |||||
| " like :sp <tab> | |||||
| set wildmenu | |||||
| " sets the size of the status bar at bottom to have a height of two | |||||
| set laststatus=2 | |||||
| " Searching when in command mode type /words to find | |||||
| " search as characters are entered | |||||
| set incsearch | |||||
| " highlight matched characters | |||||
| set hlsearch | |||||
| " Ignore case when searching | |||||
| set ignorecase | |||||
| "Disable ding sound on error, flashes cursor instead | |||||
| set visualbell | |||||
| " Display ruler on bottom right -- should be there by default | |||||
| set ruler | |||||
| " Enables mouse support | |||||
| set mouse=a | |||||
| " Auto updates file if an external source edits the file | |||||
| set autoread | |||||
| " Improves performance by only redrawing screen when needed | |||||
| set lazyredraw | |||||
| " Copy and paste | |||||
| " Selection | |||||
| " v and arrows select characters | |||||
| " V select entire lines | |||||
| " d on something selected cuts it -- also used for delete | |||||
| " y = yank = copy | |||||
| " P paste before cursor | |||||
| " p paste after cursor | |||||
| " Basic Vim navigation | |||||
| " :sp file -- this will open a new file horizontally | |||||
| " :vsp file -- will open a file splitting vertically | |||||
| " ctrl-w w -- this will toggle to another open vim window | |||||
+ 58
- 0
dfs.py
View File
| @ -0,0 +1,58 @@ | |||||
| # Python3 program to print DFS traversal | |||||
| from collections import defaultdict | |||||
| # This class represents a directed graph using | |||||
| # adjacency list representation | |||||
| class Graph: | |||||
| # Constructor | |||||
| def __init__(self): | |||||
| # default dictionary to store graph | |||||
| self.graph = defaultdict(list) | |||||
| def addEdge(self, u, v): | |||||
| self.graph[u].append(v) | |||||
| # A function used by DFS | |||||
| def DFSUtil(self, v, visited): | |||||
| # Mark the current node as visited | |||||
| # and print it | |||||
| visited[v] = True | |||||
| print(v, end = ' ') | |||||
| # Recur for all the vertices | |||||
| # adjacent to this vertex | |||||
| for i in self.graph[v]: | |||||
| if visited[i] == False: | |||||
| self.DFSUtil(i, visited) | |||||
| # The function to do DFS traversal. It uses | |||||
| # recursive DFSUtil() | |||||
| def DFS(self, v): | |||||
| # Mark all the vertices as not visited | |||||
| visited = [False] * (len(self.graph)) | |||||
| # Call the recursive helper function | |||||
| # to print DFS traversal | |||||
| self.DFSUtil(v, visited) | |||||
| # Driver code | |||||
| # Create a graph given | |||||
| # in the above diagram | |||||
| g = Graph() | |||||
| g.addEdge(0, 1) | |||||
| g.addEdge(0, 2) | |||||
| g.addEdge(1, 2) | |||||
| g.addEdge(2, 0) | |||||
| g.addEdge(2, 3) | |||||
| g.addEdge(3, 3) | |||||
| print("Following is DFS from (starting from vertex 2)") | |||||
| g.DFS(2) | |||||
+ 579
- 0
geneticAlgorithm/geneticAlgo.html
View File
| @ -0,0 +1,579 @@ | |||||
| <html> | |||||
| <head> | |||||
| <script type="text/javascript" src="https://www.gstatic.com/charts/loader.js"></script> | |||||
| <script | |||||
| src="https://code.jquery.com/jquery-3.3.1.slim.min.js" | |||||
| integrity="sha256-3edrmyuQ0w65f8gfBsqowzjJe2iM6n0nKciPUp8y+7E=" | |||||
| crossorigin="anonymous"> | |||||
| </script> | |||||
| <link href="https://stackpath.bootstrapcdn.com/bootstrap/4.3.1/css/bootstrap.min.css" rel="stylesheet" integrity="sha384-ggOyR0iXCbMQv3Xipma34MD+dH/1fQ784/j6cY/iJTQUOhcWr7x9JvoRxT2MZw1T" crossorigin="anonymous"> | |||||
| <script> | |||||
| class Gene | |||||
| { | |||||
| /** | |||||
| * Constructs a new Gene to store in a chromosome. | |||||
| * @param min minimum value that this gene can store | |||||
| * @param max value this gene can possibly be | |||||
| * @param value normalized value | |||||
| */ | |||||
| constructor(min, max, value) | |||||
| { | |||||
| this.min = min; | |||||
| this.max = max; | |||||
| this.value = value; | |||||
| } | |||||
| /** | |||||
| * De-normalizes the value of the gene | |||||
| * @returns {*} | |||||
| */ | |||||
| getRealValue() | |||||
| { | |||||
| return (this.max - this.min) * this.value + this.min; | |||||
| } | |||||
| getValue() | |||||
| { | |||||
| return this.value; | |||||
| } | |||||
| setValue(val) | |||||
| { | |||||
| this.value = val; | |||||
| } | |||||
| makeClone() | |||||
| { | |||||
| return new Gene(this.min, this.max, this.value); | |||||
| } | |||||
| makeRandomGene() | |||||
| { | |||||
| return new Gene(this.min, this.max, Math.random()); | |||||
| } | |||||
| } | |||||
| class Chromosome | |||||
| { | |||||
| /** | |||||
| * Constructs a chromosome by making a copy of | |||||
| * a list of genes. | |||||
| * @param geneArray | |||||
| */ | |||||
| constructor(geneArray) | |||||
| { | |||||
| this.genes = []; | |||||
| for(let i = 0; i < geneArray.length; i++) | |||||
| { | |||||
| this.genes.push(geneArray[i].makeClone()); | |||||
| } | |||||
| } | |||||
| getGenes() | |||||
| { | |||||
| return this.genes; | |||||
| } | |||||
| /** | |||||
| * Mutates a random gene. | |||||
| */ | |||||
| mutate() | |||||
| { | |||||
| this.genes[Math.round(Math.random() * (this.genes.length-1))].setValue(Math.random()); | |||||
| } | |||||
| /** | |||||
| * Creates a totally new chromosome with same | |||||
| * genetic structure as this chromosome but different | |||||
| * values. | |||||
| * @returns {Chromosome} | |||||
| */ | |||||
| createRandomChromosome() | |||||
| { | |||||
| let geneAr = []; | |||||
| for(let i = 0; i < this.genes.length; i++) | |||||
| { | |||||
| geneAr.push(this.genes[i].makeRandomGene()); | |||||
| } | |||||
| return new Chromosome(geneAr); | |||||
| } | |||||
| } | |||||
| /** | |||||
| * Mates two chromosomes using the blending method | |||||
| * and returns a list of 2 offspring. | |||||
| * @param father | |||||
| * @param mother | |||||
| * @returns {Chromosome[]} | |||||
| */ | |||||
| const breed = function(father, mother) | |||||
| { | |||||
| let son = new Chromosome(father.getGenes()); | |||||
| let daughter = new Chromosome(mother.getGenes()); | |||||
| for(let i = 0;i < son.getGenes().length; i++) | |||||
| { | |||||
| let blendCoef = Math.random(); | |||||
| blendGene(son.getGenes()[i], daughter.getGenes()[i], blendCoef); | |||||
| } | |||||
| return [son, daughter]; | |||||
| }; | |||||
| /** | |||||
| * Blends two genes together based on a random blend | |||||
| * coefficient. | |||||
| **/ | |||||
| const blendGene = function(gene1, gene2, blendCoef) | |||||
| { | |||||
| let value1 = (blendCoef * gene1.getValue()) + | |||||
| (gene2.getValue() * (1- blendCoef)); | |||||
| let value2 = ((1-blendCoef) * gene1.getValue()) + | |||||
| (gene2.getValue() * blendCoef); | |||||
| gene1.setValue(value1); | |||||
| gene2.setValue(value2); | |||||
| }; | |||||
| /** | |||||
| * Helper function to sort an array | |||||
| * | |||||
| * @param prop name of JSON property to sort by | |||||
| * @returns {function(*, *): number} | |||||
| */ | |||||
| function predicateBy(prop) | |||||
| { | |||||
| return function(a,b) | |||||
| { | |||||
| var result; | |||||
| if(a[prop] > b[prop]) | |||||
| { | |||||
| result = 1; | |||||
| } | |||||
| else if(a[prop] < b[prop]) | |||||
| { | |||||
| result = -1; | |||||
| } | |||||
| return result; | |||||
| } | |||||
| } | |||||
| /** | |||||
| * Function which computes the fitness of everyone in the | |||||
| * population and returns the most fit survivors. Method | |||||
| * known as elitism. | |||||
| * | |||||
| * @param population | |||||
| * @param keepNumber | |||||
| * @param fitnessFunction | |||||
| * @returns {{average: number, | |||||
| * survivors: Array, bestFit: Chromosome }} | |||||
| */ | |||||
| const naturalSelection = function(population, keepNumber, fitnessFunction) | |||||
| { | |||||
| let fitnessArray = []; | |||||
| let total = 0; | |||||
| for(let i = 0; i < population.length; i++) | |||||
| { | |||||
| const fitness = fitnessFunction(population[i]); | |||||
| console.log(fitness); | |||||
| fitnessArray.push({fit:fitness, chrom: population[i]}); | |||||
| total+= fitness; | |||||
| } | |||||
| fitnessArray.sort(predicateBy("fit")); | |||||
| let survivors = []; | |||||
| let bestFitness = fitnessArray[0].fit; | |||||
| let bestChromosome = fitnessArray[0].chrom; | |||||
| for(let i = 0; i < keepNumber; i++) | |||||
| { | |||||
| survivors.push(fitnessArray[i].chrom); | |||||
| } | |||||
| return {average: total/population.length, survivors: survivors, bestFit: bestFitness, bestChrom: bestChromosome}; | |||||
| }; | |||||
| /** | |||||
| * Randomly everyone in the population | |||||
| * | |||||
| * @param population | |||||
| * @param desiredPopulationSize | |||||
| */ | |||||
| const matePopulation = function(population, desiredPopulationSize) | |||||
| { | |||||
| const originalLength = population.length; | |||||
| while(population.length < desiredPopulationSize) | |||||
| { | |||||
| let index1 = Math.round(Math.random() * (originalLength-1)); | |||||
| let index2 = Math.round(Math.random() * (originalLength-1)); | |||||
| if(index1 !== index2) | |||||
| { | |||||
| const babies = breed(population[index1], population[index2]); | |||||
| population.push(babies[0]); | |||||
| population.push(babies[1]); | |||||
| } | |||||
| } | |||||
| }; | |||||
| /** | |||||
| * Randomly mutates the population | |||||
| **/ | |||||
| const mutatePopulation = function(population, mutatePercentage) | |||||
| { | |||||
| if(population.length >= 2) | |||||
| { | |||||
| let mutations = mutatePercentage * | |||||
| population.length * | |||||
| population[0].getGenes().length; | |||||
| for(let i = 0; i < mutations; i++) | |||||
| { | |||||
| population[i].mutate(); | |||||
| } | |||||
| } | |||||
| else | |||||
| { | |||||
| console.log("Error, population too small to mutate"); | |||||
| } | |||||
| }; | |||||
| /** | |||||
| * Introduces x random chromosomes to the population. | |||||
| * @param population | |||||
| * @param immigrationSize | |||||
| */ | |||||
| const newBlood = function(population, immigrationSize) | |||||
| { | |||||
| for(let i = 0; i < immigrationSize; i++) | |||||
| { | |||||
| let geneticChromosome = population[0]; | |||||
| population.push(geneticChromosome.createRandomChromosome()); | |||||
| } | |||||
| }; | |||||
| let costx = Math.random() * 10; | |||||
| let costy = Math.random() * 10; | |||||
| /** Defines the cost as the "distance" to a 2-d point. | |||||
| * @param chromosome | |||||
| * @returns {number} | |||||
| */ | |||||
| const basicCostFunction = function(chromosome) | |||||
| { | |||||
| return Math.abs(chromosome.getGenes()[0].getRealValue() - costx) + | |||||
| Math.abs(chromosome.getGenes()[1].getRealValue() - costy); | |||||
| }; | |||||
| /** | |||||
| * Creates a totally random population based on a desired size | |||||
| * and a prototypical chromosome. | |||||
| * | |||||
| * @param geneticChromosome | |||||
| * @param populationSize | |||||
| * @returns {Array} | |||||
| */ | |||||
| const createRandomPopulation = function(geneticChromosome, populationSize) | |||||
| { | |||||
| let population = []; | |||||
| for(let i = 0; i < populationSize; i++) | |||||
| { | |||||
| population.push(geneticChromosome.createRandomChromosome()); | |||||
| } | |||||
| return population; | |||||
| }; | |||||
| /** | |||||
| * Runs the genetic algorithm by going through the processes of | |||||
| * natural selection, mutation, mating, and immigrations. This | |||||
| * process will continue until an adequately performing chromosome | |||||
| * is found or a generation threshold is passed. | |||||
| * | |||||
| * @param geneticChromosome Prototypical chromosome: used so algo knows | |||||
| * what the dna of the population looks like. | |||||
| * @param costFunction Function which defines how bad a Chromosome is | |||||
| * @param populationSize Desired population size for population | |||||
| * @param maxGenerations Cut off level for number of generations to run | |||||
| * @param desiredCost Sufficient cost to terminate program at | |||||
| * @param mutationRate Number between [0,1] representing proportion of genes | |||||
| * to mutate each generation | |||||
| * @param keepNumber Number of Organisms which survive each generation | |||||
| * @param newBloodNumber Number of random immigrants to introduce into | |||||
| * the population each generation. | |||||
| * @returns {*} | |||||
| */ | |||||
| const runGeneticOptimization = function(geneticChromosome, costFunction, | |||||
| populationSize, maxGenerations, | |||||
| desiredCost, mutationRate, keepNumber, | |||||
| newBloodNumber) | |||||
| { | |||||
| let population = createRandomPopulation(geneticChromosome, populationSize); | |||||
| let generation = 0; | |||||
| let bestCost = Number.MAX_VALUE; | |||||
| let bestChromosome = geneticChromosome; | |||||
| do | |||||
| { | |||||
| matePopulation(population, populationSize); | |||||
| newBlood(population, newBloodNumber); | |||||
| mutatePopulation(population, mutationRate); | |||||
| let generationResult = naturalSelection(population, keepNumber, costFunction); | |||||
| if(bestCost > generationResult.bestFit) | |||||
| { | |||||
| bestChromosome = generationResult.bestChrom; | |||||
| bestCost = generationResult.bestFit; | |||||
| } | |||||
| population = generationResult.survivors; | |||||
| generation++; | |||||
| console.log("Generation " + generation + " Best Cost: " + bestCost); | |||||
| }while(generation < maxGenerations && bestCost > desiredCost); | |||||
| return bestChromosome; | |||||
| }; | |||||
| /** | |||||
| * Ugly globals used to keep track of population state for the graph. | |||||
| */ | |||||
| let genericChromosomeG, costFunctionG, | |||||
| populationSizeG, maxGenerationsG, | |||||
| desiredCostG, mutationRateG, keepNumberG, | |||||
| newBloodNumberG, populationG, generationG, | |||||
| bestCostG = Number.MAX_VALUE, bestChromosomeG = genericChromosomeG; | |||||
| const runGeneticOptimizationForGraph = function() | |||||
| { | |||||
| let generationResult = naturalSelection(populationG, keepNumberG, costFunctionG); | |||||
| stats.push([generationG, generationResult.bestFit, generationResult.average]); | |||||
| if(bestCostG > generationResult.bestFit) | |||||
| { | |||||
| bestChromosomeG = generationResult.bestChrom; | |||||
| bestCostG = generationResult.bestFit; | |||||
| } | |||||
| populationG = generationResult.survivors; | |||||
| generationG++; | |||||
| console.log("Generation " + generationG + " Best Cost: " + bestCostG); | |||||
| console.log(generationResult); | |||||
| matePopulation(populationG, populationSizeG); | |||||
| newBlood(populationG, newBloodNumberG); | |||||
| mutatePopulation(populationG, mutationRateG); | |||||
| createGraph(); | |||||
| }; | |||||
| let stats = []; | |||||
| const createGraph = function() | |||||
| { | |||||
| var dataPoints = []; | |||||
| console.log(dataPoints); | |||||
| var data = new google.visualization.DataTable(); | |||||
| data.addColumn('number', 'Gene 1'); | |||||
| data.addColumn('number', 'Gene 2'); | |||||
| for(let i = 0; i < populationG.length; i++) | |||||
| { | |||||
| data.addRow([populationG[i].getGenes()[0].getRealValue(), | |||||
| populationG[i].getGenes()[1].getRealValue()]); | |||||
| } | |||||
| var options = { | |||||
| title: 'Genetic Evolution On Two Genes Generation: ' + generationG, | |||||
| hAxis: {title: 'Gene 1', minValue: 0, maxValue: 10}, | |||||
| vAxis: {title: 'Gene 2', minValue: 0, maxValue: 10}, | |||||
| }; | |||||
| var chart = new google.visualization.ScatterChart(document.getElementById('chart_div')); | |||||
| chart.draw(data, options); | |||||
| //line chart stuff | |||||
| var line_data = new google.visualization.DataTable(); | |||||
| line_data.addColumn('number', 'Generation'); | |||||
| line_data.addColumn('number', 'Best'); | |||||
| line_data.addColumn('number', 'Average'); | |||||
| line_data.addRows(stats); | |||||
| console.log(stats); | |||||
| var lineChartOptions = { | |||||
| hAxis: { | |||||
| title: 'Generation' | |||||
| }, | |||||
| vAxis: { | |||||
| title: 'Cost' | |||||
| }, | |||||
| colors: ['#AB0D06', '#007329'] | |||||
| }; | |||||
| var chart = new google.visualization.LineChart(document.getElementById('line_chart')); | |||||
| chart.draw(line_data, lineChartOptions); | |||||
| }; | |||||
| let gene1 = new Gene(1,10,10); | |||||
| let gene2 = new Gene(1,10,0.4); | |||||
| let geneList = [gene1, gene2]; | |||||
| let exampleOrganism = new Chromosome(geneList); | |||||
| genericChromosomeG = exampleOrganism; | |||||
| costFunctionG = basicCostFunction; | |||||
| populationSizeG = 100; | |||||
| maxGenerationsG = 30; | |||||
| desiredCostG = 0.00001; | |||||
| mutationRateG = 0.3; | |||||
| keepNumberG = 30; | |||||
| newBloodNumberG = 10; | |||||
| generationG = 0; | |||||
| function verifyForm() | |||||
| { | |||||
| if(Number($("#populationSize").val()) <= 1) | |||||
| { | |||||
| alert("Population size must be greater than one."); | |||||
| return false; | |||||
| } | |||||
| if(Number($("#mutationRate").val()) > 1 || | |||||
| Number($("#mutationRate").val()) < 0) | |||||
| { | |||||
| alert("Mutation rate must be between zero and one."); | |||||
| return false; | |||||
| } | |||||
| if(Number($("#survivalSize").val()) < 0) | |||||
| { | |||||
| alert("Survival size can't be less than one."); | |||||
| return false; | |||||
| } | |||||
| if(Number($("#newBlood").val()) < 0) | |||||
| { | |||||
| alert("New organisms can't be a negative number."); | |||||
| return false; | |||||
| } | |||||
| return true; | |||||
| } | |||||
| function resetPopulation() | |||||
| { | |||||
| if(verifyForm()) | |||||
| { | |||||
| stats = []; | |||||
| autoRunning = false; | |||||
| $("#runAutoOptimizer").val("Auto Run"); | |||||
| populationSizeG = $("#populationSize").val(); | |||||
| mutationRateG = $("#mutationRate").val(); | |||||
| keepNumberG = $("#survivalSize").val(); | |||||
| newBloodNumberG = $("#newBlood").val(); | |||||
| generationG = 0; | |||||
| populationG = createRandomPopulation(genericChromosomeG, populationSizeG); | |||||
| createGraph(); | |||||
| } | |||||
| } | |||||
| populationG = createRandomPopulation(genericChromosomeG, populationSizeG); | |||||
| window.onload = function (){ | |||||
| google.charts.load('current', {packages: ['corechart', 'line']}); | |||||
| google.charts.load('current', {'packages':['corechart']}).then(function() | |||||
| { | |||||
| createGraph(); | |||||
| }) | |||||
| }; | |||||
| let autoRunning = false; | |||||
| function runAutoOptimizer() | |||||
| { | |||||
| if(autoRunning === true) | |||||
| { | |||||
| runGeneticOptimizationForGraph(); | |||||
| setTimeout(runAutoOptimizer, 1000); | |||||
| } | |||||
| } | |||||
| function startStopAutoRun() | |||||
| { | |||||
| autoRunning = !autoRunning; | |||||
| if(autoRunning) | |||||
| { | |||||
| $("#runAutoOptimizer").val("Stop Auto Run"); | |||||
| } | |||||
| else | |||||
| { | |||||
| $("#runAutoOptimizer").val("Resume Auto Run"); | |||||
| } | |||||
| runAutoOptimizer(); | |||||
| } | |||||
| //runGeneticOptimization(exampleOrganism, basicCostFunction, 100, 50, 0.01, 0.3, 20, 10); | |||||
| </script> | |||||
| </head> | |||||
| <body> | |||||
| <div id="chart_div" style="width: 900px; height: 500px;"></div> | |||||
| <div id="line_chart"></div> | |||||
| <input class='btn btn-primary' id="runOptimizer" onclick='runGeneticOptimizationForGraph()' type="button" value="Next Generation"> | |||||
| <input class='btn btn-primary' id="runAutoOptimizer" onclick='startStopAutoRun()' type="button" value="Auto Run"> | |||||
| <div class="card"> | |||||
| <div class="card-header"> | |||||
| <h2>Population Variables</h2> | |||||
| </div> | |||||
| <form class="card-body"> | |||||
| <div class="row p-2"> | |||||
| <div class="col"> | |||||
| <label for="populationSize">Population Size</label> | |||||
| <input type="text" class="form-control" value="100" id="populationSize" placeholder="Population Size" required> | |||||
| </div> | |||||
| <div class="col"> | |||||
| <label for="populationSize">Survival Size</label> | |||||
| <input type="text" class="form-control" value="20" id="survivalSize" placeholder="Survival Size" required> | |||||
| </div> | |||||
| </div> | |||||
| <div class="row p-2"> | |||||
| <div class="col"> | |||||
| <label for="populationSize">Mutation Rate</label> | |||||
| <input type="text" class="form-control" value="0.03" id="mutationRate" placeholder="Mutation Rate" required> | |||||
| </div> | |||||
| <div class="col"> | |||||
| <label for="populationSize">New Organisms Per Generation</label> | |||||
| <input type="text" class="form-control" value="5" id="newBlood" placeholder="New Organisms" required> | |||||
| </div> | |||||
| </div> | |||||
| <br> | |||||
| <input class='btn btn-primary' id="reset" onclick='resetPopulation()' type="button" value="Reset Population"> | |||||
| </form> | |||||
| </div> | |||||
| </body> | |||||
| </html> | |||||
+ 92
- 0
graphing/basicGraph.html
View File
| @ -0,0 +1,92 @@ | |||||
| <!DOCTYPE HTML> | |||||
| <html> | |||||
| <head> | |||||
| <title>Graph2d | Performance</title> | |||||
| <style> | |||||
| body, html { | |||||
| font-family: arial, sans-serif; | |||||
| font-size: 11pt; | |||||
| } | |||||
| span.label { | |||||
| width:150px; | |||||
| display:inline-block; | |||||
| } | |||||
| </style> | |||||
| <!-- note: moment.js must be loaded before vis.js, else vis.js uses its embedded version of moment.js --> | |||||
| <script src="http://cdnjs.cloudflare.com/ajax/libs/moment.js/2.8.4/moment.min.js"></script> | |||||
| <script src="https://code.jquery.com/jquery-3.3.1.min.js" | |||||
| integrity="sha256-FgpCb/KJQlLNfOu91ta32o/NMZxltwRo8QtmkMRdAu8=" | |||||
| crossorigin="anonymous"> | |||||
| </script> | |||||
| <script src="./dist/vis.js"></script> | |||||
| <link href="./dist/vis-timeline-graph2d.min.css" rel="stylesheet" type="text/css" /> | |||||
| </head> | |||||
| <body> | |||||
| <br /> | |||||
| Click the button then choose a file to graph. | |||||
| <button onclick="openFile(dispFile)">Open a file</button> | |||||
| <pre id="contents"></pre> | |||||
| <div id="visualization"> | |||||
| <script type="text/javascript"> | |||||
| function dispFile(contents) | |||||
| { | |||||
| console.log(contents); | |||||
| console.log(typeof contents); | |||||
| var container = document.getElementById('visualization'); | |||||
| var items = JSON.parse(contents); | |||||
| var dataset = new vis.DataSet(items); | |||||
| var options = {}; | |||||
| var graph2d = new vis.Graph2d(container, dataset, options); | |||||
| } | |||||
| function clickElem(elem) | |||||
| { | |||||
| // Thx user1601638 on Stack Overflow (6/6/2018 - https://stackoverflow.com/questions/13405129/javascript-create-and-save-file ) | |||||
| var eventMouse = document.createEvent("MouseEvents"); | |||||
| eventMouse.initMouseEvent("click", true, false, window, 0, 0, 0, 0, 0, false, false, false, false, 0, null); | |||||
| elem.dispatchEvent(eventMouse); | |||||
| } | |||||
| function openFile(func) | |||||
| { | |||||
| readFile = function(e) | |||||
| { | |||||
| var file = e.target.files[0]; | |||||
| if (!file) | |||||
| { | |||||
| return; | |||||
| } | |||||
| var reader = new FileReader(); | |||||
| reader.onload = function(e) | |||||
| { | |||||
| var contents = e.target.result; | |||||
| fileInput.func(contents); | |||||
| document.body.removeChild(fileInput); | |||||
| } | |||||
| reader.readAsText(file); | |||||
| } | |||||
| fileInput = document.createElement("input"); | |||||
| fileInput.type='file'; | |||||
| fileInput.style.display='none'; | |||||
| fileInput.onchange=readFile; | |||||
| fileInput.func=func; | |||||
| document.body.appendChild(fileInput); | |||||
| clickElem(fileInput); | |||||
| } | |||||
| </script> | |||||
| </div> | |||||
| </body> | |||||
| </html> | |||||
+ 1
- 0
graphing/ex.json
View File
| @ -0,0 +1 @@ | |||||
| [{"x": "2014-06-1", "y": -6},{"x": "2014-06-11", "y": 0},{"x": "2014-06-12", "y": 25},{"x": "2014-06-13", "y": 30},{"x": "2014-06-14", "y": 10},{"x": "2014-06-15", "y": 15},{"x": "2014-06-16", "y": 30}] | |||||
+ 142
- 0
graphing/floyd_warshall.py
View File
| @ -0,0 +1,142 @@ | |||||
| class Graph: | |||||
| def __init__(self): | |||||
| self.vertices = {} | |||||
| def add_vertex(self, key): | |||||
| vertex = Vertex(key) | |||||
| self.vertices[key] = vertex | |||||
| def get_vertex(self, key): | |||||
| return self.vertices[key] | |||||
| def __contains__(self, key): | |||||
| return key in self.vertices | |||||
| def add_edge(self, src_key, dest_key, weight=1): | |||||
| self.vertices[src_key].add_neighbour(self.vertices[dest_key], weight) | |||||
| def does_edge_exist(self, src_key, dest_key): | |||||
| return self.vertices[src_key].does_it_point_to(self.vertices[dest_key]) | |||||
| def __len__(self): | |||||
| return len(self.vertices) | |||||
| def __iter__(self): | |||||
| return iter(self.vertices.values()) | |||||
| class Vertex: | |||||
| def __init__(self, key): | |||||
| self.key = key | |||||
| self.points_to = {} | |||||
| def get_key(self): | |||||
| return self.key | |||||
| def add_neighbour(self, dest, weight): | |||||
| self.points_to[dest] = weight | |||||
| def get_neighbours(self): | |||||
| return self.points_to.keys() | |||||
| def get_weight(self, dest): | |||||
| return self.points_to[dest] | |||||
| def does_it_point_to(self, dest): | |||||
| return dest in self.points_to | |||||
| def floyd_warshall(g): | |||||
| distance = {v:dict.fromkeys(g, float('inf')) for v in g} | |||||
| next_v = {v:dict.fromkeys(g, None) for v in g} | |||||
| for v in g: | |||||
| for n in v.get_neighbours(): | |||||
| distance[v][n] = v.get_weight(n) | |||||
| next_v[v][n] = n | |||||
| for v in g: | |||||
| distance[v][v] = 0 | |||||
| next_v[v][v] = None | |||||
| for p in g: | |||||
| for v in g: | |||||
| for w in g: | |||||
| if distance[v][w] > distance[v][p] + distance[p][w]: | |||||
| distance[v][w] = distance[v][p] + distance[p][w] | |||||
| next_v[v][w] = next_v[v][p] | |||||
| return distance, next_v | |||||
| def print_path(next_v, u, v): | |||||
| p = u | |||||
| while (next_v[p][v]): | |||||
| print('{} -> '.format(p.get_key()), end='') | |||||
| p = next_v[p][v] | |||||
| print('{} '.format(v.get_key()), end='') | |||||
| g = Graph() | |||||
| print('Menu') | |||||
| print('add vertex <key>') | |||||
| print('add edge <src> <dest> <weight>') | |||||
| print('floyd-warshall') | |||||
| print('display') | |||||
| print('quit') | |||||
| while True: | |||||
| do = input('What would you like to do? ').split() | |||||
| operation = do[0] | |||||
| if operation == 'add': | |||||
| suboperation = do[1] | |||||
| if suboperation == 'vertex': | |||||
| key = int(do[2]) | |||||
| if key not in g: | |||||
| g.add_vertex(key) | |||||
| else: | |||||
| print('Vertex already exists.') | |||||
| elif suboperation == 'edge': | |||||
| src = int(do[2]) | |||||
| dest = int(do[3]) | |||||
| weight = int(do[4]) | |||||
| if src not in g: | |||||
| print('Vertex {} does not exist.'.format(src)) | |||||
| elif dest not in g: | |||||
| print('Vertex {} does not exist.'.format(dest)) | |||||
| else: | |||||
| if not g.does_edge_exist(src, dest): | |||||
| g.add_edge(src, dest, weight) | |||||
| else: | |||||
| print('Edge already exists.') | |||||
| elif operation == 'floyd-warshall': | |||||
| distance, next_v = floyd_warshall(g) | |||||
| print('Shortest distances:') | |||||
| for start in g: | |||||
| for end in g: | |||||
| if next_v[start][end]: | |||||
| print('From {} to {}: '.format(start.get_key(), | |||||
| end.get_key()), | |||||
| end = '') | |||||
| print_path(next_v, start, end) | |||||
| print('(distance {})'.format(distance[start][end])) | |||||
| elif operation == 'display': | |||||
| print('Vertices: ', end='') | |||||
| for v in g: | |||||
| print(v.get_key(), end=' ') | |||||
| print() | |||||
| print('Edges: ') | |||||
| for v in g: | |||||
| for dest in v.get_neighbours(): | |||||
| w = v.get_weight(dest) | |||||
| print('(src={}, dest={}, weight={}) '.format(v.get_key(), | |||||
| dest.get_key(), w)) | |||||
| print() | |||||
| elif operation == 'quit': | |||||
| break | |||||
+ 122
- 0
graphing/graph.py
View File
| @ -0,0 +1,122 @@ | |||||
| """ | |||||
| :Author: James Sherratt | |||||
| :Date: 20/10/2019 | |||||
| :License: MIT | |||||
| :name: graph.py | |||||
| """ | |||||
| class Graph: | |||||
| def __init__(self): | |||||
| self.vertices = set() | |||||
| self.edges = set() | |||||
| def add_vertex(self, vert): | |||||
| """ | |||||
| Add a vertex to the graph. | |||||
| :param vert: name of the vertex. | |||||
| :return: None | |||||
| """ | |||||
| self.vertices.add(vert) | |||||
| def add_edge(self, vert1, vert2, directional=False): | |||||
| """ | |||||
| Add an edge to the graph. The edge will be defined as a simple tuple where: | |||||
| - The first value is the initial edge. | |||||
| - The second is the final edge. | |||||
| - The third is whether the edge is directional. | |||||
| :param vert1: the start vertex. | |||||
| :param vert2: the end vertex. | |||||
| :param directional: whether or not the edge has a direction. Default: False | |||||
| :return: None | |||||
| """ | |||||
| self.vertices.add(vert1) | |||||
| self.vertices.add(vert2) | |||||
| if (not directional) and (vert1 > vert2): | |||||
| # swap if not directional to avoid duplicates in the set. | |||||
| self.edges.add((vert2, vert1, directional)) | |||||
| else: | |||||
| self.edges.add((vert1, vert2, directional)) | |||||
| def adjacency(self, vert1, vert2): | |||||
| """ | |||||
| Check if 2 vertices are adjacent (if they exist). Note: if vert1 and vert2 | |||||
| are connected, but directionally from vert2 to vert1, False is returned. | |||||
| :param vert1: The first vertex to compare. | |||||
| :param vert2: The second vertex to compare. | |||||
| :return: True if adjacent, False if not, None if either are not in the set. | |||||
| """ | |||||
| if (vert1 not in self.vertices) or (vert2 not in self.vertices): | |||||
| return None | |||||
| for edge in self.edges: | |||||
| if (vert1 == edge[0]) and (vert2 == edge[1]): | |||||
| return True | |||||
| if (not edge[2]) and ((vert1 == edge[1]) and (vert2 == edge[0])): | |||||
| return True | |||||
| return False | |||||
| def neighbours(self, vert): | |||||
| """ | |||||
| Get the neighbours of a vertex. | |||||
| :param vert: name of the vertex. | |||||
| :return: list of neighbours, or None if the vertex is not in the graph. | |||||
| """ | |||||
| if vert not in self.vertices: | |||||
| return None | |||||
| neighbours = set() | |||||
| for edge in self.edges: | |||||
| if vert == edge[0]: | |||||
| neighbours.add(edge[1]) | |||||
| elif (not edge[2]) and (vert == edge[1]): | |||||
| neighbours.add(edge[0]) | |||||
| return neighbours | |||||
| def as_dict(self): | |||||
| """ | |||||
| Convert the graph to a dictionary where: | |||||
| - Each key is a vertex. | |||||
| - Each value is a set of neighbours you can travel to. | |||||
| :return: dict representing the graph. | |||||
| """ | |||||
| graph_dict = {v: set() for v in self.vertices} | |||||
| for edge in self.edges: | |||||
| graph_dict[edge[0]].add(edge[1]) | |||||
| if not edge[2]: | |||||
| graph_dict[edge[1]].add(edge[0]) | |||||
| return graph_dict | |||||
| if __name__ == "__main__": | |||||
| mygraph = Graph() | |||||
| mygraph.add_edge("a", "b") | |||||
| mygraph.add_edge("b", "c") | |||||
| mygraph.add_edge("b", "d") | |||||
| mygraph.add_edge("c", "b") | |||||
| mygraph.add_edge("a", "e", directional=True) | |||||
| mygraph.add_vertex("z") | |||||
| print("b neighbours:", mygraph.neighbours("b")) | |||||
| print("a neighbours:", mygraph.neighbours("a")) | |||||
| print("q neighbours:", mygraph.neighbours("q")) | |||||
| print("e neighbours:", mygraph.neighbours("e")) | |||||
| print() | |||||
| # Adjacency has direction. | |||||
| print("a and e adjacent:", mygraph.adjacency("a", "e")) | |||||
| print("e and a adjacent:", mygraph.adjacency("e", "a")) | |||||
| print("d and b adjacent", mygraph.adjacency("d", "b")) | |||||
| print("q and a adjacent:", mygraph.adjacency("q", "a")) | |||||
| print("z and a adjacent:", mygraph.adjacency("z", "a")) | |||||
| print() | |||||
| print("as dict") | |||||
| print(mygraph.as_dict()) | |||||
| # Exercise/ project: add a method "path" to find path between 2 vertices. | |||||
| # (Hint: lookup DFS/ BFS algorithm.) | |||||
+ 71
- 0
multiThreadedFileIO/DataCreator.java
View File
| @ -0,0 +1,71 @@ | |||||
| import java.io.BufferedWriter; | |||||
| import java.io.File; | |||||
| import java.io.FileWriter; | |||||
| import java.math.*; | |||||
| /** | |||||
| * Simple class to generate random data for | |||||
| * file IO tests. | |||||
| * | |||||
| * @author Jeffery Russell 1-31-19 | |||||
| */ | |||||
| public class DataCreator | |||||
| { | |||||
| /** | |||||
| * Generates an obscure random character | |||||
| * @return | |||||
| */ | |||||
| private static char rndChar() | |||||
| { | |||||
| // or use Random or whatever | |||||
| int rnd = (int) (Math.random() * 52); | |||||
| char base = (rnd < 26) ? 'A' : 'a'; | |||||
| return (char) (base + rnd % 26); | |||||
| } | |||||
| /** | |||||
| * Simple function to save contents to the disk | |||||
| * | |||||
| * @param s | |||||
| * @param fileName | |||||
| */ | |||||
| private static void saveToDisk(String s, String fileName) | |||||
| { | |||||
| BufferedWriter writer; | |||||
| try | |||||
| { | |||||
| File file = new File(fileName); | |||||
| file.createNewFile(); | |||||
| writer = new BufferedWriter(new FileWriter(file)); | |||||
| writer.write(s); | |||||
| writer.flush(); | |||||
| writer.close(); | |||||
| } | |||||
| catch (Exception e) | |||||
| { | |||||
| e.printStackTrace(); | |||||
| } | |||||
| } | |||||
| /** | |||||
| * Creates 5MB of random test data to use | |||||
| */ | |||||
| public static void main(String[] arguments) | |||||
| { | |||||
| System.out.println("Creating Test Files"); | |||||
| for(int i = 0; i < 100; i++) | |||||
| { | |||||
| //10k random characters for each file | |||||
| //each file is 10kb * 500 files, total size of 5MB | |||||
| String s = ""; | |||||
| for(int j = 0; j < 1000000; j++) | |||||
| { | |||||
| s = s + rndChar(); | |||||
| } | |||||
| saveToDisk(s, "./testData/" + i + ".txt"); | |||||
| System.out.println(s); | |||||
| } | |||||
| } | |||||
| } | |||||
+ 44
- 0
multiThreadedFileIO/FileReader.java
View File
| @ -0,0 +1,44 @@ | |||||
| import java.io.BufferedReader; | |||||
| import java.io.FileInputStream; | |||||
| import java.io.IOException; | |||||
| import java.io.InputStreamReader; | |||||
| /** | |||||
| * Simple class for reading files as strings. | |||||
| * | |||||
| * @author Jeffery Russell | |||||
| */ | |||||
| public class FileReader | |||||
| { | |||||
| public static String readFile(String filePath) | |||||
| { | |||||
| String fileContent = new String(); | |||||
| try | |||||
| { | |||||
| BufferedReader br = new BufferedReader( | |||||
| new InputStreamReader(new FileInputStream(filePath))); | |||||
| String line; | |||||
| while ((line = br.readLine()) != null) | |||||
| { | |||||
| fileContent = fileContent.concat(line); | |||||
| } | |||||
| br.close(); | |||||
| } | |||||
| catch (IOException e) | |||||
| { | |||||
| e.printStackTrace(); | |||||
| } | |||||
| return fileContent; | |||||
| } | |||||
| public static void main(String[] args) | |||||
| { | |||||
| System.out.println("Test"); | |||||
| System.out.println(FileReader.readFile("./testData/data1.txt")); | |||||
| } | |||||
| } | |||||
+ 40
- 0
multiThreadedFileIO/MultiThreadedFileReadTest.java
View File
| @ -0,0 +1,40 @@ | |||||
| import java.util.*; | |||||
| /** | |||||
| * File to test the performance of multi threaded file | |||||
| * io by reading a large quantity of files in parallel | |||||
| * using a different amount of threads. | |||||
| * | |||||
| * @author Jeffery Russell 1-31-19 | |||||
| */ | |||||
| public class MultiThreadedFileReadTest | |||||
| { | |||||
| public static void main(String[] args) | |||||
| { | |||||
| List<Integer> x = new ArrayList<>(); | |||||
| List<Double> y = new ArrayList<>(); | |||||
| for(int i = 1; i <= 64; i++) | |||||
| { | |||||
| long threadTotal = 0; | |||||
| for(int w = 0; w < 20; w++) | |||||
| { | |||||
| TaskManager boss = new TaskManager(i); | |||||
| for(int j = 0; j < 500; j++) | |||||
| { | |||||
| boss.addTask(new ReadTask("./testData/" + i + ".txt")); | |||||
| } | |||||
| long startTime = System.nanoTime(); | |||||
| boss.runTasks(); | |||||
| long endTime = System.nanoTime(); | |||||
| long durationMS = (endTime - startTime)/1000000; | |||||
| threadTotal+= durationMS; | |||||
| } | |||||
| x.add(i); | |||||
| y.add(threadTotal/20.0); //finds average | |||||
| } | |||||
| System.out.println(x); | |||||
| System.out.println(y); | |||||
| } | |||||
| } | |||||
+ 20
- 0
multiThreadedFileIO/ReadTask.java
View File
| @ -0,0 +1,20 @@ | |||||
| /** | |||||
| * Simple method to be used by the task manager to do | |||||
| * file io. | |||||
| * | |||||
| * @author Jeffery Russell | |||||
| */ | |||||
| public class ReadTask | |||||
| { | |||||
| private String fileName; | |||||
| public ReadTask(String fileName) | |||||
| { | |||||
| this.fileName = fileName; | |||||
| } | |||||
| public void runTask() | |||||
| { | |||||
| FileReader.readFile(fileName); | |||||
| } | |||||
| } | |||||
+ 87
- 0
multiThreadedFileIO/TaskManager.java
View File
| @ -0,0 +1,87 @@ | |||||
| import java.util.List; | |||||
| import java.util.Vector; | |||||
| /** | |||||
| * A class which enables user to run a large chunk of | |||||
| * tasks in parallel efficiently. | |||||
| * | |||||
| * @author Jeffery 1-29-19 | |||||
| */ | |||||
| public class TaskManager | |||||
| { | |||||
| /** Number of threads to use at once */ | |||||
| private int threadCount; | |||||
| /** Meaningless tasks to run in parallel */ | |||||
| private List<ReadTask> tasks; | |||||
| public TaskManager(int threadCount) | |||||
| { | |||||
| this.threadCount = threadCount; | |||||
| //using vectors because they are thread safe | |||||
| this.tasks = new Vector<>(); | |||||
| } | |||||
| public void addTask(ReadTask t) | |||||
| { | |||||
| tasks.add(t); | |||||
| } | |||||
| /** | |||||
| * This is the fun method. | |||||
| * | |||||
| * This will run all of the tasks in parallel using the | |||||
| * desired amount of threads untill all of the jobs are | |||||
| * complete. | |||||
| */ | |||||
| public void runTasks() | |||||
| { | |||||
| int desiredThreads = threadCount > tasks.size() ? | |||||
| tasks.size() : threadCount; | |||||
| Thread[] runners = new Thread[desiredThreads]; | |||||
| for(int i = 0; i < desiredThreads; i++) | |||||
| { | |||||
| runners[i] = new Thread(()-> | |||||
| { | |||||
| ReadTask t = null; | |||||
| while(true) | |||||
| { | |||||
| //need synchronized block to prevent | |||||
| //race condition | |||||
| synchronized (tasks) | |||||
| { | |||||
| if(!tasks.isEmpty()) | |||||
| t = tasks.remove(0); | |||||
| } | |||||
| if(t == null) | |||||
| { | |||||
| break; | |||||
| } | |||||
| else | |||||
| { | |||||
| t.runTask(); | |||||
| t = null; | |||||
| } | |||||
| } | |||||
| }); | |||||
| runners[i].start(); | |||||
| } | |||||
| for(int i = 0; i < desiredThreads; i++) | |||||
| { | |||||
| try | |||||
| { | |||||
| runners[i].join(); | |||||
| } | |||||
| catch (Exception e) | |||||
| { | |||||
| e.printStackTrace(); | |||||
| } | |||||
| } | |||||
| } | |||||
| } | |||||
+ 13
- 0
multiThreadedFileIO/graph.py
View File
| @ -0,0 +1,13 @@ | |||||
| import matplotlib.pyplot as plt | |||||
| xList = [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64] | |||||
| yList = [230.5, 124.0, 84.65, 63.9, 51.6, 44.2, 39.15, 34.9, 31.9, 28.7, 27.2, 25.05, 24.25, 22.8, 21.25, 20.45, 19.8, 19.4, 18.6, 17.45, 17.0, 16.95, 16.4, 24.15, 15.2, 15.2, 15.0, 14.3, 14.7, 14.65, 14.65, 14.75, 13.85, 15.1, 14.35, 35.75, 15.3, 14.45, 14.25, 13.65, 14.45, 14.6, 13.95, 14.65, 14.5, 14.3, 14.85, 14.15, 15.0, 16.25, 14.25, 14.4, 14.15, 14.45, 14.25, 14.85, 14.3, 14.35, 15.35, 13.6, 14.3, 14.2, 14.3, 13.95] | |||||
| plt.plot(xList, yList) | |||||
| plt.xlabel('Number of Threads') | |||||
| plt.ylabel('Execution Time (MS)') | |||||
| plt.show() | |||||
+ 306
- 0
notebooks/8-bit/8-bit.ipynb
File diff suppressed because it is too large
View File
BIN
notebooks/8-bit/dolphin.jpg
View File
+ 507
- 0
notebooks/burnout/burnout.ipynb
File diff suppressed because it is too large
View File
+ 407
- 0
notebooks/cuda-vs-cpu.ipynb
File diff suppressed because it is too large
View File
+ 1168
- 0
notebooks/fitbit/GraphingMyLifeWithFitbit.ipynb
File diff suppressed because it is too large
View File
BIN
notebooks/fitbit/compression.png
View File
+ 129601
- 0
notebooks/fitbit/data/calories/calories-2019-07-01.json
File diff suppressed because it is too large
View File
+ 7486
- 0
notebooks/fitbit/data/sleep-2019-04-02.json
File diff suppressed because it is too large
View File
+ 8059
- 0
notebooks/fitbit/data/sleep-2019-06-01.json
File diff suppressed because it is too large
View File
+ 8492
- 0
notebooks/fitbit/data/sleep-2019-07-01.json
File diff suppressed because it is too large
View File
+ 6336
- 0
notebooks/fitbit/data/sleep-2019-07-31.json
File diff suppressed because it is too large
View File
+ 8372
- 0
notebooks/fitbit/data/sleep-2019-09-29.json
File diff suppressed because it is too large
View File
+ 8540
- 0
notebooks/fitbit/data/sleep-2019-11-28.json
File diff suppressed because it is too large
View File
+ 8896
- 0
notebooks/fitbit/data/sleep-2019-12-28.json
File diff suppressed because it is too large
View File
+ 182
- 0
notebooks/fitbit/data/sleep/sleep_score.csv
View File
| @ -0,0 +1,182 @@ | |||||
| sleep_log_entry_id,timestamp,overall_score,composition_score,revitalization_score,duration_score,deep_sleep_in_minutes,resting_heart_rate,restlessness | |||||
| 26093459526,2020-02-27T06:04:30Z,80,20,19,41,65,60,0.11733046286329386 | |||||
| 26081303207,2020-02-26T06:13:30Z,83,22,21,40,85,60,0.11318795430944964 | |||||
| 26062481322,2020-02-25T06:00:30Z,82,22,21,39,95,60,0.12063492063492064 | |||||
| 26045941555,2020-02-24T05:49:30Z,79,17,20,42,52,61,0.11122448979591837 | |||||
| 26034268762,2020-02-23T08:35:30Z,75,20,16,39,43,59,0.15477386934673368 | |||||
| 26022694306,2020-02-22T07:43:00Z,79,19,21,39,53,60,0.09750297265160524 | |||||
| 26007961616,2020-02-21T06:35:30Z,81,22,17,42,89,58,0.12588832487309645 | |||||
| 25988234988,2020-02-20T06:25:00Z,81,21,21,39,60,58,0.11811926605504587 | |||||
| 25969165537,2020-02-19T06:34:00Z,88,22,20,46,91,59,0.12130177514792899 | |||||
| 25952365608,2020-02-18T05:46:30Z,84,23,20,41,114,61,0.13891726251276812 | |||||
| 25938189656,2020-02-17T06:19:00Z,86,22,22,42,89,63,0.09623430962343096 | |||||
| 25926432422,2020-02-16T08:15:30Z,76,21,18,37,49,60,0.11136107986501688 | |||||
| 25901113872,2020-02-14T06:34:00Z,78,18,19,41,74,58,0.12269938650306748 | |||||
| 25883532376,2020-02-13T06:03:00Z,85,21,21,43,52,58,0.10491071428571429 | |||||
| 25871057666,2020-02-12T06:28:30Z,81,22,17,42,88,58,0.1282316442605998 | |||||
| 25860431526,2020-02-11T06:01:30Z,69,18,18,33,57,57,0.13793103448275862 | |||||
| 25844332793,2020-02-10T06:34:00Z,86,22,22,42,88,57,0.10461538461538461 | |||||
| 25829772076,2020-02-09T08:11:30Z,86,21,21,44,80,58,0.13311948676824378 | |||||
| 25817419640,2020-02-08T08:17:00Z,87,22,20,45,132,59,0.13164556962025317 | |||||
| 25803331988,2020-02-07T06:33:00Z,79,20,18,41,88,58,0.14084507042253522 | |||||
| 25785877070,2020-02-06T05:50:00Z,81,21,18,42,80,57,0.11764705882352941 | |||||
| 25769882127,2020-02-05T05:46:30Z,82,21,20,41,46,57,0.12259371833839919 | |||||
| 25755117610,2020-02-04T05:47:00Z,83,22,19,42,83,58,0.16132478632478633 | |||||
| 25742882864,2020-02-03T05:58:30Z,84,22,20,42,85,59,0.13249211356466878 | |||||
| 25729700495,2020-02-02T08:13:30Z,78,19,15,44,100,57,0.17530631479736097 | |||||
| 25717829971,2020-02-01T08:30:00Z,81,20,17,44,100,58,0.1504491017964072 | |||||
| 25704549870,2020-01-31T06:35:30Z,82,22,19,41,103,57,0.1182108626198083 | |||||
| 25684248305,2020-01-30T06:33:00Z,81,22,18,41,89,57,0.1384297520661157 | |||||
| 25655144692,2020-01-29T06:33:00Z,83,21,21,41,107,59,0.12727272727272726 | |||||
| 25642147792,2020-01-28T05:43:30Z,75,20,20,35,66,60,0.1445358401880141 | |||||
| 25624216079,2020-01-27T06:30:30Z,75,20,17,38,87,59,0.14594594594594595 | |||||
| 25613704007,2020-01-26T09:59:00Z,82,20,19,43,88,58,0.1306772908366534 | |||||
| 25593158621,2020-01-25T08:56:00Z,73,18,17,38,75,57,0.14451382694023193 | |||||
| 25584797887,2020-01-24T06:25:00Z,82,21,19,42,70,57,0.12525458248472504 | |||||
| 25566089471,2020-01-23T05:46:00Z,82,21,22,39,79,58,0.11123470522803114 | |||||
| 25553681127,2020-01-22T05:59:00Z,78,22,20,36,89,58,0.12290502793296089 | |||||
| 25536165788,2020-01-21T05:45:00Z,76,21,20,35,67,59,0.13196814562002276 | |||||
| 25523347864,2020-01-20T07:49:30Z,82,21,20,41,89,60,0.11534500514933059 | |||||
| 25510939805,2020-01-19T07:52:00Z,81,19,21,41,44,60,0.10616784630940344 | |||||
| 25498043781,2020-01-18T08:22:00Z,80,22,18,40,85,60,0.1424390243902439 | |||||
| 25481521636,2020-01-17T06:33:00Z,77,22,20,35,84,60,0.11912225705329153 | |||||
| 25464967288,2020-01-16T05:40:30Z,79,21,21,37,82,61,0.13427561837455831 | |||||
| 25451954958,2020-01-15T07:03:30Z,72,18,18,36,75,61,0.1487778958554729 | |||||
| 25433680148,2020-01-14T05:59:00Z,78,22,18,38,95,60,0.13448275862068965 | |||||
| 25416046459,2020-01-13T06:12:30Z,59,15,15,29,71,58,0.12557077625570776 | |||||
| 25404373453,2020-01-12T07:40:30Z,70,19,17,34,88,57,0.14871794871794872 | |||||
| 25391754657,2020-01-11T07:00:00Z,82,22,19,41,95,57,0.13719512195121952 | |||||
| 25376512937,2020-01-10T05:55:30Z,80,22,19,39,99,57,0.1457174638487208 | |||||
| 25363137868,2020-01-09T07:16:30Z,82,22,18,42,96,57,0.17166494312306102 | |||||
| 25342702306,2020-01-08T07:36:30Z,71,18,16,37,90,57,0.18043684710351376 | |||||
| 25320701087,2020-01-07T06:27:30Z,76,19,17,40,94,58,0.14355948869223206 | |||||
| 25302298657,2020-01-06T07:54:00Z,78,20,18,40,102,58,0.15831663326653306 | |||||
| 25290153671,2020-01-05T08:39:30Z,76,20,18,38,135,58,0.18575553416746873 | |||||
| 25277262941,2020-01-04T08:55:30Z,86,23,19,44,112,58,0.15954664341761116 | |||||
| 25260231013,2020-01-03T08:02:30Z,77,23,17,37,112,58,0.16065911431513905 | |||||
| 25246389288,2020-01-02T07:42:00Z,82,21,20,41,96,59,0.13477366255144033 | |||||
| 25230986095,2020-01-01T08:17:00Z,87,22,20,45,96,58,0.1358974358974359 | |||||
| 25223163489,2019-12-31T08:46:30Z,84,22,20,42,85,59,0.12800769971126083 | |||||
| 25208075705,2019-12-30T09:02:30Z,86,23,18,45,121,59,0.16595012897678418 | |||||
| 25192758070,2019-12-29T08:06:30Z,74,19,19,36,104,60,0.12307692307692308 | |||||
| 25179626350,2019-12-28T07:47:00Z,75,23,20,32,109,60,0.13075060532687652 | |||||
| 25167825845,2019-12-27T07:58:00Z,75,22,21,32,104,59,0.14196762141967623 | |||||
| 25152307479,2019-12-26T07:11:00Z,82,23,18,41,135,60,0.13925327951564076 | |||||
| 25141316140,2019-12-25T08:23:30Z,83,22,19,42,103,58,0.13015873015873017 | |||||
| 25130549447,2019-12-24T06:42:30Z,70,18,17,35,66,58,0.16799091940976163 | |||||
| 25119454863,2019-12-23T07:34:30Z,79,22,16,41,96,56,0.15461847389558234 | |||||
| 25108672051,2019-12-22T07:50:30Z,82,21,20,41,91,57,0.14165890027958994 | |||||
| 25098379524,2019-12-21T07:53:30Z,80,23,16,41,114,56,0.1527647610121837 | |||||
| 25089476419,2019-12-20T07:36:30Z,79,22,19,38,103,56,0.11063372717508056 | |||||
| 25072688276,2019-12-19T06:52:30Z,81,22,18,41,86,57,0.13789581205311544 | |||||
| 25057861702,2019-12-18T06:12:00Z,78,22,20,36,119,57,0.14528101802757157 | |||||
| 25043749264,2019-12-17T07:27:30Z,80,21,19,40,67,58,0.15663801337153774 | |||||
| 25030710421,2019-12-16T07:14:30Z,87,22,19,46,97,57,0.13279132791327913 | |||||
| 25017595333,2019-12-15T07:29:00Z,84,21,20,43,75,59,0.14136125654450263 | |||||
| 25007218737,2019-12-14T08:30:00Z,88,22,21,45,98,60,0.12964563526361278 | |||||
| 24997196150,2019-12-13T07:49:30Z,88,22,20,46,108,62,0.12952898550724637 | |||||
| 24982127567,2019-12-12T07:47:30Z,82,21,21,40,66,64,0.14876847290640394 | |||||
| 24972789521,2019-12-11T08:11:30Z,77,18,20,39,97,65,0.11817367949865712 | |||||
| 24958854924,2019-12-10T08:58:30Z,64,17,13,34,53,63,0.18163054695562436 | |||||
| 24944343209,2019-12-09T06:32:00Z,80,19,20,41,114,64,0.14519427402862986 | |||||
| 24933856578,2019-12-08T08:27:30Z,72,19,20,33,68,66,0.15060240963855423 | |||||
| 24924422010,2019-12-07T07:44:00Z,72,17,17,38,55,64,0.18670576735092864 | |||||
| 24911112346,2019-12-06T06:02:00Z,74,21,19,34,76,63,0.13476070528967254 | |||||
| 24901828393,2019-12-05T06:17:30Z,74,20,19,35,87,61,0.12682379349046016 | |||||
| 24889394512,2019-12-04T05:57:30Z,81,23,18,40,107,60,0.1385809312638581 | |||||
| 24866846291,2019-12-03T06:07:00Z,78,21,19,38,67,59,0.14748603351955308 | |||||
| 24854583395,2019-12-02T06:33:30Z,72,20,15,37,87,57,0.1577350859453994 | |||||
| 24845349137,2019-12-01T08:07:00Z,80,21,19,40,98,57,0.12749762131303521 | |||||
| 24832170784,2019-11-30T06:43:30Z,75,19,17,39,89,56,0.1605550049554014 | |||||
| 24823252695,2019-11-29T07:18:30Z,85,21,20,44,89,58,0.1431095406360424 | |||||
| 24810734703,2019-11-28T08:16:30Z,78,19,21,38,108,59,0.13955555555555554 | |||||
| 24800533035,2019-11-27T07:23:00Z,81,21,19,41,79,58,0.1814780168381665 | |||||
| 24789018415,2019-11-26T06:19:00Z,78,18,19,41,99,58,0.13742690058479531 | |||||
| 24775969508,2019-11-25T06:33:00Z,78,20,18,40,96,58,0.153 | |||||
| 24775969507,2019-11-24T09:13:30Z,81,23,18,40,110,57,0.14246068455134134 | |||||
| 24752259936,2019-11-23T05:36:00Z,81,18,21,42,91,59,0.1187308085977482 | |||||
| 24743421450,2019-11-22T05:40:00Z,84,19,19,46,112,60,0.14511873350923482 | |||||
| 24721924769,2019-11-21T05:39:00Z,80,20,19,41,114,60,0.1661600810536981 | |||||
| 24705802084,2019-11-20T06:03:30Z,83,18,20,45,102,61,0.15812720848056538 | |||||
| 24692804362,2019-11-19T05:59:30Z,68,19,15,34,66,59,0.232163080407701 | |||||
| 24679720563,2019-11-18T06:45:30Z,81,21,20,40,110,59,0.1457378551787351 | |||||
| 24668519678,2019-11-17T07:16:30Z,71,18,18,35,93,58,0.1624874623871615 | |||||
| 24661537014,2019-11-16T07:34:30Z,82,21,19,42,97,59,0.14965312190287414 | |||||
| 24645635181,2019-11-15T06:37:00Z,79,18,22,39,83,60,0.12270531400966184 | |||||
| 24632260941,2019-11-14T06:10:30Z,63,17,18,28,66,58,0.13878080415045396 | |||||
| 24622178551,2019-11-13T06:46:00Z,81,22,19,40,98,58,0.16093117408906882 | |||||
| 24607642569,2019-11-12T06:02:00Z,76,18,21,37,103,58,0.1277330264672037 | |||||
| 24594911463,2019-11-11T06:48:30Z,80,21,20,39,82,59,0.14729458917835672 | |||||
| 24583710526,2019-11-10T07:27:30Z,83,21,21,41,113,59,0.10933081998114987 | |||||
| 24573188107,2019-11-09T08:19:00Z,83,23,19,41,111,59,0.14476190476190476 | |||||
| 24561443781,2019-11-08T06:33:00Z,83,21,20,42,70,60,0.1506276150627615 | |||||
| 24548001288,2019-11-07T06:30:30Z,75,20,20,35,88,60,0.1248513674197384 | |||||
| 24536346513,2019-11-06T06:32:00Z,80,22,17,41,84,59,0.16270218839200762 | |||||
| 24525485244,2019-11-05T05:59:30Z,79,22,19,38,84,60,0.12485681557846506 | |||||
| 24511902336,2019-11-04T06:55:30Z,80,21,18,41,107,60,0.1648675171736997 | |||||
| 24496648737,2019-11-03T06:37:30Z,61,16,15,30,77,58,0.204739336492891 | |||||
| 24489140894,2019-11-02T08:16:30Z,87,22,20,45,96,59,0.13217866909753875 | |||||
| 24475953124,2019-11-01T06:45:00Z,81,22,22,37,94,60,0.10235294117647059 | |||||
| 24463734399,2019-10-31T06:14:00Z,41,17,13,11,75,58,0.14906832298136646 | |||||
| 24449968899,2019-10-30T06:39:30Z,82,22,19,41,94,58,0.13818860877684408 | |||||
| 24437579612,2019-10-29T06:22:30Z,76,19,21,36,73,58,0.13719185423365488 | |||||
| 24424386780,2019-10-28T06:44:30Z,84,21,21,42,90,59,0.11209439528023599 | |||||
| 24413032462,2019-10-27T06:49:30Z,69,21,14,34,71,56,0.15770609318996415 | |||||
| 24402887048,2019-10-26T05:37:00Z,75,21,19,35,93,56,0.11956521739130435 | |||||
| 24390995549,2019-10-25T06:45:30Z,81,20,19,42,95,56,0.12195121951219512 | |||||
| 24377443892,2019-10-24T07:12:30Z,75,22,18,35,83,56,0.1470292044310171 | |||||
| 24366941680,2019-10-23T06:35:30Z,84,22,22,40,113,58,0.1330603889457523 | |||||
| 24355822433,2019-10-22T06:45:30Z,82,23,18,41,110,58,0.13891726251276812 | |||||
| 24341953249,2019-10-21T06:47:00Z,79,18,20,41,65,57,0.13636363636363635 | |||||
| 24341953248,2019-10-20T07:45:30Z,85,20,21,44,110,56,0.11251049538203191 | |||||
| 24318195646,2019-10-19T08:19:30Z,79,21,20,38,86,56,0.1321266968325792 | |||||
| 24306066115,2019-10-18T07:12:00Z,84,24,20,40,127,56,0.11683168316831684 | |||||
| 24289622902,2019-10-17T06:36:30Z,75,21,20,34,69,56,0.15337423312883436 | |||||
| 24278460408,2019-10-16T06:21:00Z,79,21,21,37,117,57,0.10526315789473684 | |||||
| 24269469092,2019-10-15T07:59:00Z,84,22,21,41,93,58,0.11545988258317025 | |||||
| 24250954876,2019-10-14T05:13:00Z,76,19,20,37,77,58,0.1293833131801693 | |||||
| 24242836711,2019-10-13T09:18:30Z,63,21,14,28,60,56,0.15173237753882915 | |||||
| 24235096825,2019-10-12T08:10:00Z,82,22,20,40,107,57,0.1335914811229429 | |||||
| 24219310077,2019-10-11T06:49:30Z,71,20,19,32,97,58,0.1372328458942632 | |||||
| 24207395605,2019-10-10T06:22:00Z,71,20,16,35,74,57,0.14365256124721604 | |||||
| 24192906384,2019-10-09T06:52:00Z,83,21,22,40,81,58,0.12016293279022404 | |||||
| 24178673308,2019-10-08T06:19:30Z,81,20,21,40,91,59,0.11836283185840708 | |||||
| 24157324429,2019-10-07T07:33:30Z,77,19,19,39,78,59,0.17323556370302476 | |||||
| 24138872551,2019-10-06T09:19:30Z,78,21,16,41,134,60,0.23254139668826493 | |||||
| 24131744325,2019-10-05T06:37:30Z,80,22,20,38,98,60,0.1513903192584964 | |||||
| 24118453273,2019-10-04T06:23:30Z,81,22,19,40,92,60,0.13224043715846995 | |||||
| 24107294203,2019-10-03T06:11:00Z,76,19,21,36,77,61,0.14412136536030343 | |||||
| 24094261384,2019-10-02T06:49:30Z,80,22,19,39,125,61,0.155053974484789 | |||||
| 24081125459,2019-10-01T06:36:00Z,79,21,20,38,73,60,0.12017167381974249 | |||||
| 24067314952,2019-09-30T06:08:30Z,76,20,19,37,51,59,0.14870689655172414 | |||||
| 24054803851,2019-09-29T07:21:00Z,78,22,19,37,92,59,0.1380065717415115 | |||||
| 24043257064,2019-09-28T07:00:30Z,66,18,19,29,86,58,0.15037593984962405 | |||||
| 24032032616,2019-09-27T06:47:30Z,85,22,21,42,101,59,0.11288711288711288 | |||||
| 24018795264,2019-09-26T06:36:00Z,73,18,17,38,92,58,0.12582781456953643 | |||||
| 24008259982,2019-09-25T07:42:00Z,82,20,18,44,77,58,0.15384615384615385 | |||||
| 23995153240,2019-09-24T06:20:30Z,83,22,20,41,98,58,0.13559322033898305 | |||||
| 23983598772,2019-09-23T06:47:00Z,81,20,20,41,108,59,0.143796992481203 | |||||
| 23973042208,2019-09-22T07:31:00Z,77,19,17,41,101,58,0.16177957532861476 | |||||
| 23973042207,2019-09-21T07:15:00Z,79,19,20,40,124,57,0.1359126984126984 | |||||
| 23947975535,2019-09-20T06:42:00Z,86,20,21,45,117,59,0.11261261261261261 | |||||
| 23933467238,2019-09-19T06:19:00Z,79,17,17,45,85,57,0.14273356401384082 | |||||
| 23910315823,2019-09-17T05:13:30Z,66,17,19,30,98,56,0.16644113667117727 | |||||
| 23891736899,2019-09-16T06:40:00Z,84,21,21,42,80,57,0.12987012987012986 | |||||
| 23880011727,2019-09-15T07:25:30Z,77,20,18,39,89,57,0.1579466929911155 | |||||
| 23869387147,2019-09-14T07:18:00Z,70,17,18,35,87,56,0.14772727272727273 | |||||
| 23857222270,2019-09-13T06:42:00Z,70,21,17,32,128,55,0.12357217030114226 | |||||
| 23843106439,2019-09-12T06:29:30Z,78,22,18,38,85,54,0.15171650055370986 | |||||
| 23828701593,2019-09-11T05:59:00Z,81,22,20,39,86,55,0.1271186440677966 | |||||
| 23813261926,2019-09-10T06:18:30Z,77,21,19,37,68,55,0.13847900113507378 | |||||
| 23800207430,2019-09-09T06:37:00Z,81,21,20,40,78,55,0.12413108242303873 | |||||
| 23779847904,2019-09-08T07:02:30Z,85,23,20,42,117,56,0.13186813186813187 | |||||
| 23767509651,2019-09-07T07:03:00Z,75,21,19,35,77,55,0.11543287327478043 | |||||
| 23753447582,2019-09-06T06:41:30Z,79,21,18,40,91,56,0.15695952615992104 | |||||
| 23738011029,2019-09-05T06:13:00Z,78,21,21,36,80,57,0.1261574074074074 | |||||
| 23724233662,2019-09-04T06:43:30Z,82,22,19,41,95,56,0.13779128672745694 | |||||
| 23709362778,2019-09-03T06:31:00Z,85,23,20,42,115,55,0.12966804979253113 | |||||
| 23696231032,2019-09-02T07:38:30Z,79,20,20,39,88,56,0.17092337917485265 | |||||
| 23684345925,2019-09-01T07:15:30Z,84,22,21,41,95,56,0.1332675222112537 | |||||
| 23673204871,2019-08-31T07:11:00Z,74,18,21,35,73,56,0.10270270270270271 | |||||
| 23661278483,2019-08-30T06:34:00Z,73,17,19,37,50,55,0.12108559498956159 | |||||
| 23646265400,2019-08-29T05:55:00Z,80,21,21,38,61,57,0.11296076099881094 | |||||
+ 118294
- 0
notebooks/fitbit/data/steps-2019-04-02.json
File diff suppressed because it is too large
View File
+ 124426
- 0
notebooks/fitbit/data/steps-2019-05-02.json
File diff suppressed because it is too large
View File
+ 122410
- 0
notebooks/fitbit/data/steps-2019-06-01.json
File diff suppressed because it is too large
View File
+ 123349
- 0
notebooks/fitbit/data/steps-2019-07-01.json
File diff suppressed because it is too large
View File
+ 93460
- 0
notebooks/fitbit/data/steps-2019-07-31.json
File diff suppressed because it is too large
View File
+ 126109
- 0
notebooks/fitbit/data/steps-2019-08-30.json
File diff suppressed because it is too large
View File
+ 125791
- 0
notebooks/fitbit/data/steps-2019-09-29.json
File diff suppressed because it is too large
View File
+ 123265
- 0
notebooks/fitbit/data/steps-2019-10-29.json
File diff suppressed because it is too large
View File
+ 121957
- 0
notebooks/fitbit/data/steps-2019-11-28.json
File diff suppressed because it is too large
View File
+ 124729
- 0
notebooks/fitbit/data/steps-2019-12-28.json
File diff suppressed because it is too large
View File
+ 123559
- 0
notebooks/fitbit/data/steps-2020-01-27.json
File diff suppressed because it is too large
View File
+ 5272
- 0
notebooks/fitbit/data/steps-2020-02-26.json
File diff suppressed because it is too large
View File
BIN
notebooks/fitbit/dataExport.png
View File
+ 428
- 0
notebooks/fitbit/fitbitsleep.ipynb
File diff suppressed because it is too large
View File
BIN
notebooks/fitbit/restingHeartRate.jpg
View File
+ 1124
- 0
notebooks/fitbit/restingHeartRate.svg
File diff suppressed because it is too large
View File
BIN
notebooks/k-means/bg.jpg
View File
BIN
notebooks/k-means/filters.png
View File
+ 587
- 0
notebooks/k-means/k-means.ipynb
File diff suppressed because it is too large
View File
+ 43
- 0
notebooks/node_2_vec/friendsMap.map
View File
| @ -0,0 +1,43 @@ | |||||
| 76561198188400721 jrtechs | |||||
| 76561198049526995 Noosh | |||||
| 76561198067517157 nweis76 | |||||
| 76561198131175960 Delta_∆ | |||||
| 76561198084464357 Ceta | |||||
| 76561198085584420 PureMaths | |||||
| 76561198068647768 Void Cause | |||||
| 76561198094968588 Dumcumpster | |||||
| 76561198107069713 yankeeman1081 | |||||
| 76561198086854442 jspike397 | |||||
| 76561198273751201 Kyon | |||||
| 76561198083124245 drichardson005 | |||||
| 76561198035606013 BK🐻 | |||||
| 76561198255835078 Zelazny | |||||
| 76561198170096391 Greata | |||||
| 76561198306786411 fazeboojujee | |||||
| 76561198062501319 Cantankerous | |||||
| 76561198036270560 Toaster | |||||
| 76561198306443796 numstudequals1 | |||||
| 76561198133044936 Panda | |||||
| 76561198095814450 Tbonedog | |||||
| 76561198069739846 Alminikar | |||||
| 76561198233398192 Saxophones of Death™ | |||||
| 76561198162654610 Raydan | |||||
| 76561198047367972 Oberyn Martell | |||||
| 76561198057450983 TraceTheKitteh | |||||
| 76561198176504246 shistthis | |||||
| 76561198111538799 sergiozygmunt | |||||
| 76561198853827591 theramendragon | |||||
| 76561198055948417 Xaldin31 | |||||
| 76561198137304077 lilsar419 | |||||
| 76561198406334664 wywyit | |||||
| 76561198098761042 wee haw | |||||
| 76561198192510666 Rebel | |||||
| 76561198066260593 NUT | |||||
| 76561198099625445 DAW Legacy | |||||
| 76561198017734545 Which Ones Pink? | |||||
| 76561198036176189 Ben | |||||
| 76561198069739485 Rkames517 | |||||
| 76561198062300654 Rawls | |||||
| 76561198068098265 Brendy Flannel | |||||
| 76561198065642391 Therefore | |||||
| 76561198121369685 DataFrogman | |||||
+ 320
- 0
notebooks/node_2_vec/jrtechs.edgelist
View File
| @ -0,0 +1,320 @@ | |||||
| 76561198035606013 76561198069739846 | |||||
| 76561198035606013 76561198065642391 | |||||
| 76561198035606013 76561198057450983 | |||||
| 76561198035606013 76561198131175960 | |||||
| 76561198035606013 76561198094968588 | |||||
| 76561198035606013 76561198036176189 | |||||
| 76561198084464357 76561198036270560 | |||||
| 76561198084464357 76561198017734545 | |||||
| 76561198084464357 76561198065642391 | |||||
| 76561198084464357 76561198133044936 | |||||
| 76561198084464357 76561198085584420 | |||||
| 76561198084464357 76561198306786411 | |||||
| 76561198084464357 76561198111538799 | |||||
| 76561198084464357 76561198057450983 | |||||
| 76561198084464357 76561198255835078 | |||||
| 76561198084464357 76561198069739485 | |||||
| 76561198084464357 76561198176504246 | |||||
| 76561198084464357 76561198086854442 | |||||
| 76561198084464357 76561198107069713 | |||||
| 76561198084464357 76561198406334664 | |||||
| 76561198084464357 76561198069739846 | |||||
| 76561198084464357 76561198068098265 | |||||
| 76561198084464357 76561198062300654 | |||||
| 76561198084464357 76561198131175960 | |||||
| 76561198084464357 76561198094968588 | |||||
| 76561198084464357 76561198036176189 | |||||
| 76561198036270560 76561198084464357 | |||||
| 76561198036270560 76561198095814450 | |||||
| 76561198036270560 76561198062300654 | |||||
| 76561198036270560 76561198049526995 | |||||
| 76561198036270560 76561198057450983 | |||||
| 76561198036270560 76561198131175960 | |||||
| 76561198036270560 76561198255835078 | |||||
| 76561198121369685 76561198068098265 | |||||
| 76561198121369685 76561198192510666 | |||||
| 76561198121369685 76561198085584420 | |||||
| 76561198121369685 76561198162654610 | |||||
| 76561198121369685 76561198086854442 | |||||
| 76561198065642391 76561198035606013 | |||||
| 76561198065642391 76561198084464357 | |||||
| 76561198065642391 76561198017734545 | |||||
| 76561198065642391 76561198133044936 | |||||
| 76561198065642391 76561198306786411 | |||||
| 76561198065642391 76561198095814450 | |||||
| 76561198065642391 76561198057450983 | |||||
| 76561198065642391 76561198069739485 | |||||
| 76561198065642391 76561198176504246 | |||||
| 76561198065642391 76561198069739846 | |||||
| 76561198065642391 76561198137304077 | |||||
| 76561198065642391 76561198062300654 | |||||
| 76561198065642391 76561198049526995 | |||||
| 76561198065642391 76561198083124245 | |||||
| 76561198065642391 76561198047367972 | |||||
| 76561198065642391 76561198131175960 | |||||
| 76561198065642391 76561198094968588 | |||||
| 76561198065642391 76561198036176189 | |||||
| 76561198085584420 76561198107069713 | |||||
| 76561198085584420 76561198084464357 | |||||
| 76561198085584420 76561198406334664 | |||||
| 76561198085584420 76561198068098265 | |||||
| 76561198085584420 76561198121369685 | |||||
| 76561198085584420 76561198192510666 | |||||
| 76561198085584420 76561198162654610 | |||||
| 76561198085584420 76561198086854442 | |||||
| 76561198095814450 76561198036270560 | |||||
| 76561198095814450 76561198065642391 | |||||
| 76561198095814450 76561198133044936 | |||||
| 76561198095814450 76561198306786411 | |||||
| 76561198095814450 76561198099625445 | |||||
| 76561198095814450 76561198057450983 | |||||
| 76561198095814450 76561198069739485 | |||||
| 76561198095814450 76561198255835078 | |||||
| 76561198095814450 76561198069739846 | |||||
| 76561198095814450 76561198062300654 | |||||
| 76561198095814450 76561198049526995 | |||||
| 76561198095814450 76561198047367972 | |||||
| 76561198095814450 76561198131175960 | |||||
| 76561198095814450 76561198036176189 | |||||
| 76561198111538799 76561198107069713 | |||||
| 76561198111538799 76561198084464357 | |||||
| 76561198111538799 76561198406334664 | |||||
| 76561198111538799 76561198068098265 | |||||
| 76561198111538799 76561198192510666 | |||||
| 76561198111538799 76561198162654610 | |||||
| 76561198111538799 76561198086854442 | |||||
| 76561198057450983 76561198035606013 | |||||
| 76561198057450983 76561198084464357 | |||||
| 76561198057450983 76561198036270560 | |||||
| 76561198057450983 76561198017734545 | |||||
| 76561198057450983 76561198098761042 | |||||
| 76561198057450983 76561198065642391 | |||||
| 76561198057450983 76561198133044936 | |||||
| 76561198057450983 76561198306786411 | |||||
| 76561198057450983 76561198095814450 | |||||
| 76561198057450983 76561198255835078 | |||||
| 76561198057450983 76561198069739485 | |||||
| 76561198057450983 76561198176504246 | |||||
| 76561198057450983 76561198069739846 | |||||
| 76561198057450983 76561198137304077 | |||||
| 76561198057450983 76561198062300654 | |||||
| 76561198057450983 76561198049526995 | |||||
| 76561198057450983 76561198306443796 | |||||
| 76561198057450983 76561198036176189 | |||||
| 76561198099625445 76561198069739846 | |||||
| 76561198099625445 76561198095814450 | |||||
| 76561198099625445 76561198094968588 | |||||
| 76561198099625445 76561198176504246 | |||||
| 76561198255835078 76561198084464357 | |||||
| 76561198255835078 76561198036270560 | |||||
| 76561198255835078 76561198069739846 | |||||
| 76561198255835078 76561198017734545 | |||||
| 76561198255835078 76561198095814450 | |||||
| 76561198255835078 76561198062300654 | |||||
| 76561198255835078 76561198057450983 | |||||
| 76561198255835078 76561198131175960 | |||||
| 76561198233398192 76561198137304077 | |||||
| 76561198233398192 76561198066260593 | |||||
| 76561198233398192 76561198133044936 | |||||
| 76561198233398192 76561198170096391 | |||||
| 76561198055948417 76561198306786411 | |||||
| 76561198086854442 76561198107069713 | |||||
| 76561198086854442 76561198084464357 | |||||
| 76561198086854442 76561198406334664 | |||||
| 76561198086854442 76561198068098265 | |||||
| 76561198086854442 76561198121369685 | |||||
| 76561198086854442 76561198192510666 | |||||
| 76561198086854442 76561198085584420 | |||||
| 76561198086854442 76561198111538799 | |||||
| 76561198176504246 76561198084464357 | |||||
| 76561198176504246 76561198065642391 | |||||
| 76561198176504246 76561198306786411 | |||||
| 76561198176504246 76561198099625445 | |||||
| 76561198176504246 76561198057450983 | |||||
| 76561198176504246 76561198131175960 | |||||
| 76561198107069713 76561198084464357 | |||||
| 76561198107069713 76561198068098265 | |||||
| 76561198107069713 76561198192510666 | |||||
| 76561198107069713 76561198085584420 | |||||
| 76561198107069713 76561198111538799 | |||||
| 76561198107069713 76561198162654610 | |||||
| 76561198107069713 76561198086854442 | |||||
| 76561198406334664 76561198084464357 | |||||
| 76561198406334664 76561198068098265 | |||||
| 76561198406334664 76561198085584420 | |||||
| 76561198406334664 76561198111538799 | |||||
| 76561198406334664 76561198086854442 | |||||
| 76561198137304077 76561198066260593 | |||||
| 76561198137304077 76561198065642391 | |||||
| 76561198137304077 76561198133044936 | |||||
| 76561198137304077 76561198062300654 | |||||
| 76561198137304077 76561198047367972 | |||||
| 76561198137304077 76561198057450983 | |||||
| 76561198137304077 76561198170096391 | |||||
| 76561198137304077 76561198233398192 | |||||
| 76561198062300654 76561198084464357 | |||||
| 76561198062300654 76561198036270560 | |||||
| 76561198062300654 76561198017734545 | |||||
| 76561198062300654 76561198065642391 | |||||
| 76561198062300654 76561198133044936 | |||||
| 76561198062300654 76561198095814450 | |||||
| 76561198062300654 76561198057450983 | |||||
| 76561198062300654 76561198255835078 | |||||
| 76561198062300654 76561198069739485 | |||||
| 76561198062300654 76561198069739846 | |||||
| 76561198062300654 76561198137304077 | |||||
| 76561198062300654 76561198049526995 | |||||
| 76561198062300654 76561198131175960 | |||||
| 76561198062300654 76561198094968588 | |||||
| 76561198062300654 76561198036176189 | |||||
| 76561198083124245 76561198065642391 | |||||
| 76561198083124245 76561198036176189 | |||||
| 76561198047367972 76561198017734545 | |||||
| 76561198047367972 76561198137304077 | |||||
| 76561198047367972 76561198065642391 | |||||
| 76561198047367972 76561198133044936 | |||||
| 76561198047367972 76561198095814450 | |||||
| 76561198047367972 76561198049526995 | |||||
| 76561198047367972 76561198131175960 | |||||
| 76561198047367972 76561198069739485 | |||||
| 76561198094968588 76561198035606013 | |||||
| 76561198094968588 76561198084464357 | |||||
| 76561198094968588 76561198069739846 | |||||
| 76561198094968588 76561198017734545 | |||||
| 76561198094968588 76561198065642391 | |||||
| 76561198094968588 76561198062300654 | |||||
| 76561198094968588 76561198099625445 | |||||
| 76561198094968588 76561198036176189 | |||||
| 76561198170096391 76561198137304077 | |||||
| 76561198170096391 76561198066260593 | |||||
| 76561198170096391 76561198133044936 | |||||
| 76561198170096391 76561198233398192 | |||||
| 76561198036176189 76561198035606013 | |||||
| 76561198036176189 76561198084464357 | |||||
| 76561198036176189 76561198069739846 | |||||
| 76561198036176189 76561198098761042 | |||||
| 76561198036176189 76561198065642391 | |||||
| 76561198036176189 76561198133044936 | |||||
| 76561198036176189 76561198095814450 | |||||
| 76561198036176189 76561198062300654 | |||||
| 76561198036176189 76561198083124245 | |||||
| 76561198036176189 76561198057450983 | |||||
| 76561198036176189 76561198094968588 | |||||
| 76561198036176189 76561198069739485 | |||||
| 76561198017734545 76561198084464357 | |||||
| 76561198017734545 76561198069739846 | |||||
| 76561198017734545 76561198133044936 | |||||
| 76561198017734545 76561198098761042 | |||||
| 76561198017734545 76561198065642391 | |||||
| 76561198017734545 76561198062300654 | |||||
| 76561198017734545 76561198049526995 | |||||
| 76561198017734545 76561198047367972 | |||||
| 76561198017734545 76561198057450983 | |||||
| 76561198017734545 76561198094968588 | |||||
| 76561198017734545 76561198255835078 | |||||
| 76561198017734545 76561198069739485 | |||||
| 76561198133044936 76561198084464357 | |||||
| 76561198133044936 76561198017734545 | |||||
| 76561198133044936 76561198066260593 | |||||
| 76561198133044936 76561198065642391 | |||||
| 76561198133044936 76561198095814450 | |||||
| 76561198133044936 76561198057450983 | |||||
| 76561198133044936 76561198233398192 | |||||
| 76561198133044936 76561198069739485 | |||||
| 76561198133044936 76561198062501319 | |||||
| 76561198133044936 76561198069739846 | |||||
| 76561198133044936 76561198137304077 | |||||
| 76561198133044936 76561198062300654 | |||||
| 76561198133044936 76561198049526995 | |||||
| 76561198133044936 76561198067517157 | |||||
| 76561198133044936 76561198047367972 | |||||
| 76561198133044936 76561198170096391 | |||||
| 76561198133044936 76561198036176189 | |||||
| 76561198098761042 76561198017734545 | |||||
| 76561198098761042 76561198057450983 | |||||
| 76561198098761042 76561198306443796 | |||||
| 76561198098761042 76561198036176189 | |||||
| 76561198066260593 76561198133044936 | |||||
| 76561198066260593 76561198137304077 | |||||
| 76561198066260593 76561198170096391 | |||||
| 76561198066260593 76561198233398192 | |||||
| 76561198306786411 76561198084464357 | |||||
| 76561198306786411 76561198069739846 | |||||
| 76561198306786411 76561198065642391 | |||||
| 76561198306786411 76561198095814450 | |||||
| 76561198306786411 76561198057450983 | |||||
| 76561198306786411 76561198131175960 | |||||
| 76561198306786411 76561198055948417 | |||||
| 76561198306786411 76561198176504246 | |||||
| 76561198162654610 76561198107069713 | |||||
| 76561198162654610 76561198121369685 | |||||
| 76561198162654610 76561198192510666 | |||||
| 76561198162654610 76561198085584420 | |||||
| 76561198162654610 76561198111538799 | |||||
| 76561198069739485 76561198084464357 | |||||
| 76561198069739485 76561198017734545 | |||||
| 76561198069739485 76561198069739846 | |||||
| 76561198069739485 76561198065642391 | |||||
| 76561198069739485 76561198133044936 | |||||
| 76561198069739485 76561198095814450 | |||||
| 76561198069739485 76561198062300654 | |||||
| 76561198069739485 76561198049526995 | |||||
| 76561198069739485 76561198047367972 | |||||
| 76561198069739485 76561198057450983 | |||||
| 76561198069739485 76561198131175960 | |||||
| 76561198069739485 76561198036176189 | |||||
| 76561198062501319 76561198133044936 | |||||
| 76561198069739846 76561198035606013 | |||||
| 76561198069739846 76561198084464357 | |||||
| 76561198069739846 76561198017734545 | |||||
| 76561198069739846 76561198065642391 | |||||
| 76561198069739846 76561198133044936 | |||||
| 76561198069739846 76561198306786411 | |||||
| 76561198069739846 76561198095814450 | |||||
| 76561198069739846 76561198099625445 | |||||
| 76561198069739846 76561198057450983 | |||||
| 76561198069739846 76561198069739485 | |||||
| 76561198069739846 76561198255835078 | |||||
| 76561198069739846 76561198062300654 | |||||
| 76561198069739846 76561198131175960 | |||||
| 76561198069739846 76561198094968588 | |||||
| 76561198069739846 76561198036176189 | |||||
| 76561198068098265 76561198107069713 | |||||
| 76561198068098265 76561198084464357 | |||||
| 76561198068098265 76561198406334664 | |||||
| 76561198068098265 76561198121369685 | |||||
| 76561198068098265 76561198192510666 | |||||
| 76561198068098265 76561198085584420 | |||||
| 76561198068098265 76561198111538799 | |||||
| 76561198068098265 76561198086854442 | |||||
| 76561198192510666 76561198107069713 | |||||
| 76561198192510666 76561198068098265 | |||||
| 76561198192510666 76561198121369685 | |||||
| 76561198192510666 76561198085584420 | |||||
| 76561198192510666 76561198111538799 | |||||
| 76561198192510666 76561198162654610 | |||||
| 76561198192510666 76561198086854442 | |||||
| 76561198049526995 76561198036270560 | |||||
| 76561198049526995 76561198017734545 | |||||
| 76561198049526995 76561198065642391 | |||||
| 76561198049526995 76561198133044936 | |||||
| 76561198049526995 76561198095814450 | |||||
| 76561198049526995 76561198062300654 | |||||
| 76561198049526995 76561198047367972 | |||||
| 76561198049526995 76561198057450983 | |||||
| 76561198049526995 76561198069739485 | |||||
| 76561198067517157 76561198133044936 | |||||
| 76561198131175960 76561198035606013 | |||||
| 76561198131175960 76561198084464357 | |||||
| 76561198131175960 76561198036270560 | |||||
| 76561198131175960 76561198069739846 | |||||
| 76561198131175960 76561198065642391 | |||||
| 76561198131175960 76561198306786411 | |||||
| 76561198131175960 76561198095814450 | |||||
| 76561198131175960 76561198062300654 | |||||
| 76561198131175960 76561198047367972 | |||||
| 76561198131175960 76561198255835078 | |||||
| 76561198131175960 76561198069739485 | |||||
| 76561198131175960 76561198176504246 | |||||
| 76561198306443796 76561198098761042 | |||||
| 76561198306443796 76561198057450983 | |||||
+ 39
- 0
notebooks/node_2_vec/output/jrtechs2.emd
View File
| @ -0,0 +1,39 @@ | |||||
| 76561198084464357 0.22637059 0.17309421 -0.047417082 -0.19539477 -0.376943 -0.022090694 0.12190686 0.006333136 0.17078407 -0.16434675 -0.14344274 0.0707061 0.03752186 0.09877573 -0.22592762 0.10418106 0.14492817 0.15903844 -0.18958774 -0.024045361 -0.13320665 -0.06923964 -0.12813589 -0.3280573 0.13117692 -0.015366782 -0.12967604 -0.035129875 0.01031167 -0.011651808 -0.12473403 0.074550755 -0.06524019 -0.08758856 -0.019999007 -0.022523584 -0.004174753 -0.03748225 -0.046775363 -0.013586961 -0.2888517 -0.04860712 -0.19155592 -0.0029932323 0.2037822 0.18742456 -0.24540155 0.3507938 -0.109798744 -0.10331786 | |||||
| 76561198057450983 0.18881463 0.10484256 -0.07110202 -0.24082139 -0.40187156 0.11362265 0.18980016 0.06074355 0.3114207 -0.35041252 -0.084652916 -0.03986029 -0.057815213 0.061492413 -0.12260288 0.022724794 0.15843162 0.15134184 -0.07626594 -0.10567249 -0.31308064 -0.016841307 -0.04648596 -0.18032297 0.20023072 -0.10150606 -0.183425 0.035480183 0.12023826 0.01781087 -0.08787196 0.0135735255 -0.09438313 -0.040423732 0.025793772 -0.16082627 -0.02614664 -0.043824583 -0.03541194 0.101386406 -0.26133397 0.01812578 -0.14152019 -0.016886769 0.11592071 0.14048438 -0.213555 0.26810238 -0.23459966 0.022427179 | |||||
| 76561198065642391 0.16117236 0.12707286 -0.049071565 -0.22475749 -0.38269418 0.08124416 0.16345559 0.026218422 0.24946508 -0.29118553 -0.032005124 -0.013123474 -0.037144214 0.099733986 -0.16442856 0.017743498 0.20041893 0.15649626 -0.115774125 -0.076802604 -0.273641 -0.037228003 -0.05052939 -0.20305239 0.17849866 -0.09719955 -0.15639313 0.03444168 0.1260998 -0.019039676 -0.0982309 0.04167126 -0.12563835 -0.03058053 0.0019653616 -0.16748299 -0.019394407 -0.07811248 -0.04692965 0.08688936 -0.31315535 -0.0038551528 -0.1597786 0.021446709 0.13752647 0.1296905 -0.24591756 0.2978024 -0.24371934 0.029849164 | |||||
| 76561198133044936 0.079512894 0.07041713 -0.018858409 -0.22705422 -0.407911 0.13462968 0.19161797 -0.023506047 0.3192814 -0.27165064 -0.23337689 -0.11832532 -0.07606436 0.053831678 -0.05066651 0.08262957 0.31861186 0.18440625 -0.06490549 -0.10523448 -0.28386927 0.10107621 -0.14824352 -0.13747391 0.22416355 0.052063297 -0.07368989 0.029049309 0.21563941 0.14961143 -0.14053574 0.07976514 -0.10555467 -0.09587351 -0.0548718 -0.1019019 -0.11951104 0.014595861 -0.13663863 0.020653818 -0.25344658 -0.022718936 -0.22289884 -0.048778895 0.0401405 0.12216337 -0.3172736 0.09288634 -0.070460975 0.05220961 | |||||
| 76561198062300654 0.22250521 0.11693102 -0.077717416 -0.2627228 -0.43860975 0.102072835 0.2164471 0.08948511 0.31378153 -0.39153564 -0.044383664 0.002873929 -0.037035033 0.08218612 -0.17385834 0.02304037 0.14096145 0.1758448 -0.09229181 -0.12743269 -0.30886117 -0.040116988 -0.024903556 -0.21198611 0.18670943 -0.15098059 -0.19961299 0.06701814 0.11677947 -0.029612694 -0.08474066 0.019907234 -0.09353158 -0.0077796727 0.055685442 -0.1557845 0.0004210872 -0.074875124 0.0012693922 0.12909964 -0.242743 0.020810105 -0.08904973 0.022175908 0.08994497 0.08945654 -0.1655352 0.25148526 -0.26067913 0.031106705 | |||||
| 76561198069739846 0.19302638 0.10344062 -0.05425505 -0.23925813 -0.37238845 0.10448243 0.18253765 0.0435905 0.30280897 -0.3441143 -0.0683461 -0.031187892 -0.05977858 0.076245785 -0.13491338 0.014964683 0.1530082 0.17164314 -0.06370337 -0.109781586 -0.31881398 -0.027924553 -0.05530244 -0.17358772 0.2097243 -0.121022895 -0.20274468 0.06456121 0.15158892 0.0050261877 -0.092144236 0.020480443 -0.12846431 -0.032061853 0.0320777 -0.17494796 -0.01790394 -0.062649004 -0.033561904 0.1082481 -0.30862418 0.020739578 -0.1329093 -0.009725911 0.10687202 0.109754115 -0.22186637 0.27921742 -0.26998332 0.053189274 | |||||
| 76561198095814450 0.16667746 0.091015086 -0.07347555 -0.26523334 -0.40914062 0.14942275 0.24062552 0.09428762 0.32831088 -0.3834571 -0.02174694 -0.08062243 -0.0940943 0.08172432 -0.097146705 -0.028984185 0.16745195 0.17733684 -0.04802745 -0.10639211 -0.3449221 -0.010498022 -0.034632627 -0.12991816 0.24422307 -0.14980032 -0.19625974 0.036451112 0.14554851 0.006842924 -0.06748595 0.02676372 -0.12196199 0.015600681 0.06910712 -0.22079591 -0.046598632 -0.06303662 -0.03742557 0.14238463 -0.30833668 -0.0024973517 -0.112285316 -0.0047185267 0.1459727 0.14589567 -0.20610014 0.294961 -0.288551 0.05864515 | |||||
| 76561198036176189 0.2181946 0.1456158 -0.063216165 -0.23596789 -0.44432867 0.08354943 0.22378144 0.05557387 0.29959002 -0.37387443 -0.013365661 -0.011593603 -0.06839467 0.093986765 -0.19268224 -0.020571794 0.14974935 0.1518645 -0.10864363 -0.10048686 -0.34091967 -0.03928709 -0.001949952 -0.19133992 0.19132295 -0.16518652 -0.21802539 0.0792085 0.14096105 -0.05009671 -0.111111246 0.024421113 -0.122660846 -0.032062966 0.030396989 -0.19005105 0.007291107 -0.07750875 0.018624594 0.12822498 -0.2684167 0.019973218 -0.106695384 0.008099264 0.07790111 0.06015877 -0.18536285 0.2666641 -0.29988953 0.06108308 | |||||
| 76561198017734545 0.1844068 0.10791857 -0.055086304 -0.22677076 -0.42117056 0.1191463 0.24901557 0.0691128 0.34359846 -0.38548806 -0.073591605 -0.05741242 -0.07418889 0.10757234 -0.14173773 0.0137533955 0.21219616 0.17369936 -0.079123504 -0.096924774 -0.39793384 0.011017707 -0.06763745 -0.1768744 0.2463748 -0.15990648 -0.23136435 0.07388335 0.20901404 0.0055857403 -0.124804035 0.017127799 -0.13264523 -0.030653091 0.05313412 -0.23143743 -0.043516915 -0.06925246 -0.014446211 0.10092704 -0.2313569 0.017327696 -0.060766846 -0.028390046 0.01359708 0.024991293 -0.17798783 0.22584164 -0.24291082 0.085779786 | |||||
| 76561198131175960 0.18709633 0.1159048 -0.06235683 -0.24904335 -0.41933104 0.1329422 0.23412463 0.06185049 0.3241728 -0.36808455 -0.06263462 -0.054166023 -0.0819914 0.093538426 -0.13308565 0.002593986 0.18274297 0.1613406 -0.06707555 -0.10734584 -0.34892014 -0.018364865 -0.03199263 -0.1609787 0.20114924 -0.11629508 -0.1870345 0.048710063 0.14726876 0.0050412286 -0.096454464 0.0032559463 -0.12326516 -0.014995486 0.0456062 -0.19881293 -0.03276473 -0.06482951 -0.027943596 0.114527665 -0.24728297 0.013423705 -0.10039417 -0.001298922 0.08140864 0.082277365 -0.18818747 0.23623629 -0.23898323 0.06907876 | |||||
| 76561198069739485 0.16863635 0.10223341 -0.060545985 -0.23197044 -0.41741177 0.10444429 0.2316712 0.06514049 0.30053082 -0.33041558 -0.05668571 -0.05348813 -0.054245885 0.0888209 -0.119278625 0.05207464 0.19055451 0.17565472 -0.09638077 -0.093090564 -0.3223323 0.007907617 -0.08228502 -0.19251926 0.23392947 -0.09926177 -0.18060961 0.024472993 0.14110212 0.039337307 -0.08071854 0.032271996 -0.111653715 0.0015317283 0.034901317 -0.16781302 -0.04611637 -0.038588554 -0.064283356 0.07447686 -0.24495794 0.019286042 -0.12717813 0.001665474 0.09649522 0.13303445 -0.18640113 0.24702369 -0.21232006 0.042585194 | |||||
| 76561198049526995 0.16421597 0.08703316 -0.014224388 -0.28656122 -0.4452673 0.1730569 0.24652794 0.0893594 0.33507857 -0.3432222 -0.04287437 -0.09887701 -0.08301029 0.0785178 -0.10616222 0.033054214 0.22304797 0.17388679 -0.0842754 -0.110752985 -0.37500057 0.02052552 -0.08197923 -0.16772889 0.2477338 -0.10421696 -0.18422544 0.051990423 0.17595439 0.03956827 -0.09467426 0.047493536 -0.12648419 0.016369984 0.054430306 -0.18480542 -0.054272182 -0.060079213 -0.053671844 0.106657766 -0.23742117 -0.014950159 -0.09810321 0.0019860014 0.08148887 0.08952626 -0.17329475 0.19852565 -0.20817707 0.056783915 | |||||
| 76561198086854442 0.34254107 0.30957827 -0.11524153 -0.18334594 -0.45872745 -0.16210774 0.062599085 -0.084785044 0.085214816 -0.03368019 -0.32834852 0.25391206 0.14481808 0.11519705 -0.35920364 0.29045287 0.10589501 0.12952629 -0.29110923 -0.01102346 0.08528755 -0.12225788 -0.22754893 -0.49886554 0.037407905 0.17435938 -0.011043183 -0.18952647 -0.21453218 -0.017247276 -0.24563341 0.078471065 0.010968881 -0.21681622 -0.15290259 0.26152828 0.020307522 0.054162256 -0.06659027 -0.18470995 -0.25736004 -0.024604902 -0.3297332 -0.0099238 0.4460233 0.4319575 -0.2738786 0.45831376 0.04806938 -0.38181412 | |||||
| 76561198085584420 0.32387844 0.29968905 -0.09340624 -0.17020656 -0.4588123 -0.17199877 0.09759802 -0.07764273 0.08653636 -0.04453819 -0.33883795 0.23766832 0.15137453 0.105054416 -0.36854342 0.28447026 0.12303037 0.14410329 -0.30316597 -0.010570295 0.046115037 -0.1285634 -0.22102548 -0.5085482 0.042155243 0.12994027 -0.024931712 -0.1772889 -0.20540316 -0.039178245 -0.22197098 0.081810065 0.019409591 -0.19457516 -0.11883437 0.22094813 0.027794816 0.04066784 -0.057719074 -0.16206896 -0.22455785 -0.04280631 -0.2705832 0.00254194 0.37553343 0.34916008 -0.2524561 0.41834506 0.03628819 -0.34442592 | |||||
| 76561198068098265 0.32968754 0.29224247 -0.09760475 -0.17074688 -0.4280352 -0.15662102 0.08407618 -0.07047733 0.1104908 -0.049096562 -0.30009216 0.2276259 0.13768554 0.10759053 -0.33203432 0.25383678 0.10350801 0.13466042 -0.28075787 0.010627751 0.05177611 -0.119647026 -0.21901429 -0.4779795 0.054432567 0.14214997 -0.025383443 -0.16434667 -0.18549423 -0.009664651 -0.20851217 0.07405772 0.009354706 -0.19233598 -0.110344075 0.20606421 0.018651392 0.052316774 -0.05603669 -0.15958522 -0.21974984 -0.0388657 -0.26515332 -0.0037708012 0.35297117 0.33556804 -0.23458849 0.39969453 0.05403758 -0.32730108 | |||||
| 76561198255835078 0.20422879 0.1337637 -0.055301018 -0.23732188 -0.41868728 0.08202576 0.21261728 0.07931523 0.26944122 -0.3411403 -0.003033904 -0.009268218 -0.051814515 0.07968477 -0.15937214 -0.0101328 0.13801226 0.15221834 -0.10284773 -0.09293722 -0.3116927 -0.033873197 -0.030841395 -0.19593075 0.19186153 -0.16313156 -0.18322457 0.05660241 0.11838679 -0.026428936 -0.071605235 0.030191686 -0.09954815 0.00139449 0.040899087 -0.19298099 -0.004946959 -0.091355175 -0.014682736 0.109668195 -0.26626796 0.022318676 -0.115954876 0.0015445307 0.12967266 0.14838426 -0.2004102 0.31037828 -0.25866765 0.026230749 | |||||
| 76561198094968588 0.23099363 0.15101755 -0.049652692 -0.2500952 -0.40849155 0.08924417 0.19487293 0.034900554 0.2942292 -0.36203057 -0.062242344 0.024216892 -0.048077025 0.08870414 -0.19530663 -0.01682613 0.1369601 0.16583551 -0.072400734 -0.11296437 -0.30924445 -0.06823371 -0.016871605 -0.2022507 0.17886162 -0.12835509 -0.18100967 0.06528005 0.09193228 -0.052074566 -0.11000872 0.009431187 -0.11892662 -0.038558077 0.0154938055 -0.17211698 0.003627009 -0.07355131 0.002812495 0.13359915 -0.27408114 0.024543943 -0.09876224 -0.003284637 0.10332882 0.06608704 -0.17527495 0.26352975 -0.27293444 0.01622719 | |||||
| 76561198306786411 0.23577496 0.15582335 -0.019733993 -0.26907524 -0.46246845 0.12697245 0.17553708 0.06440576 0.2933275 -0.35662764 0.089201055 0.04344786 -0.049513053 0.13977818 -0.2230863 -0.032304075 0.1711684 0.15170828 -0.11081198 -0.12706006 -0.40165755 -0.10961717 -0.049708348 -0.23554803 0.20453718 -0.20538126 -0.20086817 0.07469057 0.14608128 -0.10709947 -0.08981632 0.041590784 -0.15272382 0.013766062 0.018011538 -0.23727441 0.03338206 -0.14506945 0.03026656 0.19288088 -0.24228245 0.027329555 -0.062434774 0.01310837 0.16087131 0.09016839 -0.11262507 0.2550445 -0.2581555 0.011757311 | |||||
| 76561198137304077 0.05084607 0.06377567 0.029904949 -0.28008777 -0.40055475 0.16508777 0.18005909 -0.033626378 0.34804904 -0.24664168 -0.24834043 -0.17009543 -0.08987903 0.07188041 -0.029420521 0.10113487 0.3681934 0.18576343 -0.062915765 -0.112020835 -0.3517709 0.12343172 -0.18351123 -0.12096594 0.24584693 0.068532 -0.096090384 0.02159829 0.2517719 0.18457109 -0.16981202 0.07980096 -0.14879091 -0.08591049 -0.061774842 -0.10609378 -0.13957348 -0.0063995877 -0.14989343 0.025805552 -0.242199 -0.054443225 -0.20876794 -0.06542156 0.010888981 0.086591795 -0.3248936 0.029740417 -0.020795887 0.080167554 | |||||
| 76561198047367972 0.15275417 0.09440066 -0.046161544 -0.24256153 -0.39044595 0.10920197 0.17191173 0.01972117 0.29987305 -0.29182997 -0.157382 -0.048863977 -0.038603302 0.08367885 -0.11625347 0.07863301 0.23812206 0.19126749 -0.08724187 -0.10746165 -0.28421468 0.039807376 -0.108612075 -0.18033825 0.2031557 -0.012254289 -0.12654842 0.03660974 0.16271141 0.07516582 -0.13266632 0.05608405 -0.11699921 -0.06430921 -0.008142919 -0.09756316 -0.057883263 -0.016529424 -0.05701254 0.055081207 -0.25308317 -0.020628037 -0.15820476 -0.03139943 0.07289092 0.11251792 -0.2497786 0.17986298 -0.14975126 0.020175178 | |||||
| 76561198111538799 0.3036404 0.27613953 -0.10101969 -0.20036869 -0.47141036 -0.12553287 0.06401144 -0.119364664 0.104999974 -0.011282191 -0.41150567 0.22151628 0.12879363 0.112925746 -0.32282853 0.29434735 0.15068853 0.14617495 -0.2649083 -0.0025120883 0.04731047 -0.06054026 -0.25552198 -0.46518323 0.080555245 0.20927878 0.0035025629 -0.19554582 -0.14960007 0.028762102 -0.2655043 0.120591745 -0.0017705827 -0.22710508 -0.17538425 0.25740537 -0.0070566335 0.08758394 -0.09060272 -0.21231057 -0.22795713 -0.058392636 -0.33592728 -0.031041555 0.3482926 0.33693632 -0.29145777 0.32310814 0.11992054 -0.34634903 | |||||
| 76561198192510666 0.3474313 0.30692083 -0.10706236 -0.1990391 -0.47386932 -0.16174832 0.053934477 -0.11045854 0.103935435 -0.01870137 -0.39013398 0.2555468 0.16538395 0.09464734 -0.36461172 0.30979285 0.10817218 0.15884669 -0.3080719 0.013434673 0.0859855 -0.10265159 -0.23406328 -0.51304233 0.052208208 0.20126481 -0.013518984 -0.19859777 -0.20180504 0.002393528 -0.23610297 0.0953092 0.02466766 -0.23664194 -0.16356464 0.29505062 0.017506557 0.092672825 -0.06693318 -0.2342259 -0.22791708 -0.03342698 -0.33860508 -0.028792575 0.38162994 0.38219008 -0.28770107 0.3755938 0.095116146 -0.38313824 | |||||
| 76561198107069713 0.38418534 0.33296037 -0.12762368 -0.17852028 -0.45819336 -0.19529031 0.07528587 -0.083179004 0.10940843 -0.059054233 -0.37238896 0.2905615 0.16192137 0.108712904 -0.40243933 0.2736475 0.081959985 0.15972969 -0.31503195 -0.0012036243 0.07953631 -0.13735458 -0.21672371 -0.5389489 0.0392174 0.15120299 -0.027223136 -0.18476981 -0.22191137 -0.05411894 -0.23175688 0.07529672 0.052297886 -0.23222485 -0.110240415 0.23511553 0.035543308 0.05762436 -0.024124298 -0.16959715 -0.2048569 -0.050579876 -0.25984573 -0.0026981186 0.36232162 0.34324473 -0.21938014 0.41711733 0.057436377 -0.37593263 | |||||
| 76561198036270560 0.14397192 0.08334548 -0.04469934 -0.2561699 -0.4225368 0.14867412 0.21092238 0.058282316 0.313992 -0.35724682 -0.040337965 -0.095076576 -0.06561623 0.09265195 -0.09453865 0.045526065 0.2606633 0.16861185 -0.05913528 -0.12292606 -0.35649815 0.033481017 -0.07746854 -0.12556142 0.2294217 -0.11348743 -0.16947931 0.07688432 0.19763827 0.049573503 -0.09148916 0.03140501 -0.14623797 0.00046233187 0.02777242 -0.17530276 -0.056076273 -0.050583236 -0.06638729 0.10550991 -0.27991408 0.012155746 -0.14809363 0.016292194 0.07506183 0.112898804 -0.2201068 0.20800517 -0.22228128 0.055588257 | |||||
| 76561198035606013 0.22403184 0.13860935 -0.07170239 -0.23633002 -0.3851042 0.06962776 0.25138408 0.049882714 0.35110667 -0.39081424 -0.0784561 -0.031130273 -0.063622996 0.10138198 -0.16820188 -0.02749013 0.14189342 0.17802821 -0.08234558 -0.08815543 -0.33991873 -0.052402038 -0.01718193 -0.18273875 0.22613233 -0.13184868 -0.22050497 0.039693583 0.12279355 0.005959421 -0.09681493 -0.018152623 -0.114482865 -0.03636636 0.03285335 -0.20866942 -0.000112431924 -0.08251096 0.0045912866 0.10603749 -0.2362029 0.043653633 -0.06954407 -0.021909706 0.073642775 0.074181 -0.18845314 0.27974162 -0.2565502 0.06752613 | |||||
| 76561198176504246 0.2284575 0.14545225 -0.046393584 -0.23783854 -0.41206592 0.09284343 0.15882511 0.028858747 0.2785178 -0.31970385 -0.050608598 0.04732327 -0.03284202 0.10197963 -0.21470807 0.023948532 0.17378537 0.16852878 -0.11449056 -0.108416624 -0.288645 -0.08230517 -0.071623296 -0.23142359 0.17396913 -0.09209991 -0.13887164 0.03040408 0.06400619 -0.041824076 -0.12375758 0.041198444 -0.10426776 -0.03620996 0.008661757 -0.11931847 -0.0121125085 -0.06188938 -0.0022041074 0.12413089 -0.25194913 -6.069077e-05 -0.10160189 0.013302347 0.16848335 0.11541641 -0.15720437 0.24258807 -0.20898528 -0.041242257 | |||||
| 76561198121369685 0.36825195 0.32509533 -0.11736185 -0.17219964 -0.4916117 -0.20539656 0.05276573 -0.11258646 0.04586855 -0.00970232 -0.3958037 0.2975072 0.18550335 0.10964625 -0.41070074 0.33184403 0.10514816 0.15209007 -0.33077535 0.0050268155 0.112141736 -0.124768175 -0.24047749 -0.5607367 0.029459236 0.18100987 0.010657332 -0.22585817 -0.2490769 -0.0405968 -0.23392515 0.09480492 0.053320188 -0.24275787 -0.14380261 0.29350024 0.035643235 0.085761555 -0.05726756 -0.21882486 -0.22846258 -0.040771533 -0.32726178 0.014338213 0.40005514 0.3836112 -0.25366268 0.43122298 0.078945294 -0.40346253 | |||||
| 76561198162654610 0.34695917 0.32070547 -0.10050611 -0.17910023 -0.4568615 -0.21021856 0.08406304 -0.104268186 0.10773384 -0.029855613 -0.42233047 0.28087804 0.18237478 0.11074125 -0.38688207 0.3250932 0.14440548 0.17826994 -0.33091044 0.025190417 0.090570636 -0.113041535 -0.25421536 -0.562643 0.05482017 0.20456815 -0.008464956 -0.18983297 -0.19874062 0.004034344 -0.2416712 0.10931306 0.037552014 -0.2335336 -0.13965298 0.2631763 -0.0011963076 0.070183724 -0.091388926 -0.22647984 -0.22218983 -0.0494376 -0.3240695 -0.017113559 0.3604048 0.36381122 -0.3040433 0.39249942 0.10331354 -0.36881155 | |||||
| 76561198406334664 0.28305718 0.25869372 -0.095679 -0.18264408 -0.41270575 -0.13238613 0.06889177 -0.07670079 0.12410099 -0.067683905 -0.3341638 0.19581027 0.13485932 0.092432775 -0.31416708 0.2555328 0.11869398 0.14523543 -0.26656982 -0.019504383 0.03910543 -0.09800471 -0.22110392 -0.44035456 0.05997275 0.15466471 -0.022513755 -0.15973592 -0.16345102 0.003923755 -0.20436099 0.07741159 0.0051217885 -0.19334693 -0.12499778 0.19843556 0.008181473 0.055967722 -0.076557904 -0.15694065 -0.21828173 -0.034587182 -0.28119585 -0.013511817 0.33842176 0.35085726 -0.2671838 0.3721947 0.050504435 -0.32589602 | |||||
| 76561198098761042 0.21111481 0.124898545 -0.06675497 -0.25015622 -0.36927617 0.07900296 0.24098006 0.10832148 0.37492865 -0.4189536 -0.09497082 -0.07379196 -0.08979049 0.04720399 -0.13692069 -0.04665753 0.10380948 0.19285686 -0.08932293 -0.08939387 -0.3550219 -0.020346515 -0.03026678 -0.16322915 0.2378477 -0.15314841 -0.24570547 0.03311747 0.15484545 0.020133022 -0.096255824 -0.00013513748 -0.09627935 -0.03387609 0.08945824 -0.25980866 -0.026094073 -0.08544189 0.00920267 0.11699284 -0.24506095 0.035363454 -0.0669694 -0.082532644 0.04892704 0.10205599 -0.21944775 0.28150722 -0.27296388 0.10587777 | |||||
| 76561198170096391 0.0069788 0.005919656 0.051495407 -0.32079914 -0.4073266 0.23348217 0.171251 -0.06311195 0.373394 -0.2241381 -0.36583677 -0.24454768 -0.089249425 0.05788221 0.033699363 0.16043496 0.47002196 0.20430273 -0.04187704 -0.15235455 -0.3499455 0.20683546 -0.2217468 -0.06541704 0.2695047 0.19451709 -0.049059104 0.024842022 0.32965752 0.31723747 -0.24643245 0.111958995 -0.17807698 -0.12467712 -0.09345244 -0.034506533 -0.21199925 0.03258025 -0.21684842 -0.013332772 -0.261481 -0.07761844 -0.26663408 -0.0844894 -0.049469367 0.061394736 -0.42410803 -0.10678735 0.03989682 0.13155942 | |||||
| 76561198066260593 -0.0012319235 0.0018281628 0.023123825 -0.33053738 -0.3924185 0.2472896 0.16956325 -0.0608233 0.3843416 -0.2237018 -0.33327287 -0.23764051 -0.09602595 0.043903504 0.056503646 0.14061612 0.4795503 0.20970406 -0.050004832 -0.14200833 -0.33629456 0.21809374 -0.22140649 -0.051260374 0.30881768 0.20381816 -0.030661566 0.03181241 0.3603506 0.34539744 -0.24437547 0.12426482 -0.16791798 -0.15895678 -0.13545848 -0.0553565 -0.20505448 0.045219764 -0.22169755 -0.04982665 -0.24254869 -0.07196838 -0.29574284 -0.09731388 -0.031014172 0.08635339 -0.43166023 -0.100182384 0.07344307 0.119933665 | |||||
| 76561198233398192 0.002090749 -0.019404763 0.03552148 -0.33156437 -0.3981038 0.26195565 0.16810834 -0.040420774 0.40588227 -0.21742727 -0.37480575 -0.26947838 -0.09307771 0.045861054 0.052752934 0.14719073 0.46502012 0.22194266 -0.025898535 -0.15193766 -0.35606676 0.22763512 -0.21730317 -0.049230706 0.30872586 0.21986806 -0.06211104 0.04279583 0.37989628 0.36092278 -0.26809344 0.12603697 -0.19916567 -0.12780918 -0.106969945 -0.05540217 -0.22773463 0.03833827 -0.22044288 -0.023669915 -0.24687687 -0.1123604 -0.2749139 -0.115704335 -0.06394924 0.035009824 -0.4492996 -0.14808506 0.086609416 0.14714402 | |||||
| 76561198099625445 0.2055478 0.12837608 -0.05503023 -0.2344446 -0.4076341 0.11420073 0.21084942 0.06502717 0.30410096 -0.36516762 0.00718494 0.0005491337 -0.05003054 0.11183439 -0.16368742 -0.01161308 0.15040927 0.17278118 -0.076006725 -0.10837558 -0.35160053 -0.06839973 -0.012475045 -0.19501446 0.1951187 -0.18213958 -0.20493712 0.082270525 0.12650141 -0.051296666 -0.08864778 0.011173826 -0.12863064 -0.005714577 0.03197137 -0.18616295 0.007001383 -0.09139288 0.01735045 0.104694255 -0.26359707 0.032273732 -0.10945158 0.017646449 0.12599283 0.09992092 -0.18550509 0.29857588 -0.261369 0.013124895 | |||||
| 76561198306443796 0.20952201 0.1082174 -0.058894258 -0.22635081 -0.36429945 0.08868942 0.2145234 0.118072115 0.34663963 -0.38532454 -0.057764117 -0.09209741 -0.08521907 0.036962103 -0.13144097 -0.010477377 0.10396321 0.19054501 -0.096804455 -0.06677834 -0.34803393 -0.038097873 -0.059931688 -0.19666065 0.21909525 -0.14953011 -0.25832 0.040402032 0.1349521 0.011416193 -0.0735839 0.01511647 -0.08712464 -0.049181025 0.07137101 -0.21843548 -0.016298052 -0.07046689 0.012860036 0.114801824 -0.24479301 0.029679382 -0.086947955 -0.070472956 0.060814783 0.10588453 -0.2316168 0.31899905 -0.24149619 0.039814994 | |||||
| 76561198083124245 0.18784085 0.11630195 -0.06590356 -0.20987189 -0.37137064 0.080536194 0.22644602 0.080131985 0.337934 -0.38297108 0.0010978709 -0.041674294 -0.07443796 0.10718557 -0.16031294 -0.045074925 0.11317768 0.18544613 -0.07911185 -0.09390654 -0.35544115 -0.06825034 -0.03126699 -0.19823997 0.23052976 -0.17158622 -0.2493543 0.049892716 0.14350945 -0.005205546 -0.0673639 -0.008497373 -0.12741543 0.0050176787 0.07148565 -0.24560645 -0.007314682 -0.09385316 0.015243395 0.15861993 -0.28863758 0.036965366 -0.072681546 -0.025075842 0.08361631 0.07841974 -0.19370948 0.3200258 -0.3049125 0.07649351 | |||||
| 76561198062501319 0.06587496 0.054355495 -0.01758024 -0.28811067 -0.408295 0.2000756 0.21908408 0.005989786 0.36173987 -0.27244255 -0.25412953 -0.21298829 -0.096377134 0.07297794 -0.014620238 0.07513211 0.3740009 0.17620595 -0.0563191 -0.12347753 -0.35775837 0.124346405 -0.13421129 -0.08276982 0.26213446 0.04821704 -0.12654042 0.039581597 0.31390294 0.21367665 -0.20093554 0.0543149 -0.17631225 -0.1054289 -0.034343824 -0.13611986 -0.15922198 -0.013324192 -0.1355989 0.021675432 -0.2505345 -0.054176256 -0.1918642 -0.07296861 -0.021516832 0.0506421 -0.34379992 0.056857232 -0.09682778 0.13625515 | |||||
| 76561198055948417 0.24326533 0.2160671 -0.02706927 -0.2864412 -0.4753786 0.10273929 0.14638449 0.032030385 0.28945908 -0.32167968 0.04248631 0.0785814 -0.036810715 0.15118515 -0.24673927 -0.047023214 0.158946 0.14187415 -0.12522215 -0.11113887 -0.3256903 -0.11801299 -0.055638634 -0.24089533 0.18373406 -0.15424119 -0.14814895 -0.0064601693 0.06938204 -0.121468335 -0.103591695 0.028775329 -0.13639693 0.013498841 0.006718703 -0.19217138 0.018144151 -0.13041031 0.03014829 0.16025434 -0.24780382 -0.033504553 -0.044934288 -0.005349496 0.21337706 0.09522274 -0.07615483 0.2574353 -0.20301828 -0.036485445 | |||||
| 76561198067517157 0.045390874 0.04874821 -0.0034944667 -0.27203554 -0.38045105 0.20452732 0.22411284 -0.0021262174 0.3592525 -0.2700214 -0.26435822 -0.239218 -0.118945226 0.07518768 -0.013540715 0.06506123 0.37545067 0.19849579 -0.054887153 -0.1295746 -0.38225782 0.13126 -0.16380523 -0.08588551 0.2648634 0.06802869 -0.1167201 0.03737488 0.3099814 0.23071188 -0.19691646 0.059150826 -0.1732653 -0.09852894 -0.020030558 -0.15594837 -0.17611912 -0.011522908 -0.15991443 0.0257558 -0.25372055 -0.060826913 -0.18793182 -0.07687972 -0.007320919 0.08122364 -0.34911227 0.060074463 -0.08621058 0.13632911 | |||||
+ 278
- 0
notebooks/node_2_vec/steam_node_2_vec.ipynb
File diff suppressed because it is too large
View File
+ 288
- 0
notebooks/parallel-java.ipynb
File diff suppressed because it is too large
View File
+ 1063
- 0
notebooks/pytorch-headfirst.ipynb
File diff suppressed because it is too large
View File
+ 1
- 0
notebooks/steam/games.json
File diff suppressed because it is too large
View File
BIN
notebooks/steam/playTimes.png
View File
+ 1454
- 0
notebooks/steam/steam.ipynb
File diff suppressed because it is too large
View File
+ 486
- 0
notebooks/word-embeddings-part2/embeddings.ipynb
File diff suppressed because it is too large
View File
BIN
notebooks/word-embeddings-part2/ex1.png
View File
BIN
notebooks/word-embeddings-part2/smallex.png
View File
+ 284
- 0
notebooks/word-embeddings/Untitled.ipynb
View File
| @ -0,0 +1,284 @@ | |||||
| { | |||||
| "cells": [ | |||||
| { | |||||
| "cell_type": "code", | |||||
| "execution_count": 1, | |||||
| "metadata": {}, | |||||
| "outputs": [], | |||||
| "source": [ | |||||
| "import gensim\n" | |||||
| ] | |||||
| }, | |||||
| { | |||||
| "cell_type": "code", | |||||
| "execution_count": 4, | |||||
| "metadata": {}, | |||||
| "outputs": [], | |||||
| "source": [ | |||||
| "model = gensim.models.KeyedVectors.load_word2vec_format('./GoogleNews-vectors-negative300.bin', binary=True) " | |||||
| ] | |||||
| }, | |||||
| { | |||||
| "cell_type": "code", | |||||
| "execution_count": 6, | |||||
| "metadata": {}, | |||||
| "outputs": [ | |||||
| { | |||||
| "data": { | |||||
| "text/plain": [ | |||||
| "[('hi', 0.654898464679718),\n", | |||||
| " ('goodbye', 0.639905571937561),\n", | |||||
| " ('howdy', 0.6310957074165344),\n", | |||||
| " ('goodnight', 0.5920578241348267),\n", | |||||
| " ('greeting', 0.5855878591537476),\n", | |||||
| " ('Hello', 0.5842196941375732),\n", | |||||
| " (\"g'day\", 0.5754077434539795),\n", | |||||
| " ('See_ya', 0.5688871145248413),\n", | |||||
| " ('ya_doin', 0.5643119812011719),\n", | |||||
| " ('greet', 0.5636603832244873)]" | |||||
| ] | |||||
| }, | |||||
| "execution_count": 6, | |||||
| "metadata": {}, | |||||
| "output_type": "execute_result" | |||||
| } | |||||
| ], | |||||
| "source": [ | |||||
| "model.most_similar(\"hello\")" | |||||
| ] | |||||
| }, | |||||
| { | |||||
| "cell_type": "code", | |||||
| "execution_count": 8, | |||||
| "metadata": {}, | |||||
| "outputs": [ | |||||
| { | |||||
| "data": { | |||||
| "text/plain": [ | |||||
| "[('coders', 0.6104331612586975),\n", | |||||
| " ('coder', 0.6063331365585327),\n", | |||||
| " ('Coding', 0.5804804563522339),\n", | |||||
| " ('formatting', 0.5671651363372803),\n", | |||||
| " ('soluble_receptors', 0.5576372146606445),\n", | |||||
| " ('ICD9', 0.5571348667144775),\n", | |||||
| " ('refactoring', 0.5495434999465942),\n", | |||||
| " ('database_schemas', 0.5372464656829834),\n", | |||||
| " ('recode', 0.534299373626709),\n", | |||||
| " ('XHTML_CSS', 0.5328801870346069)]" | |||||
| ] | |||||
| }, | |||||
| "execution_count": 8, | |||||
| "metadata": {}, | |||||
| "output_type": "execute_result" | |||||
| } | |||||
| ], | |||||
| "source": [ | |||||
| "model.most_similar(\"coding\")" | |||||
| ] | |||||
| }, | |||||
| { | |||||
| "cell_type": "code", | |||||
| "execution_count": 9, | |||||
| "metadata": {}, | |||||
| "outputs": [ | |||||
| { | |||||
| "data": { | |||||
| "text/plain": [ | |||||
| "[('cats', 0.8099379539489746),\n", | |||||
| " ('dog', 0.7609456777572632),\n", | |||||
| " ('kitten', 0.7464985251426697),\n", | |||||
| " ('feline', 0.7326233983039856),\n", | |||||
| " ('beagle', 0.7150583267211914),\n", | |||||
| " ('puppy', 0.7075453996658325),\n", | |||||
| " ('pup', 0.6934291124343872),\n", | |||||
| " ('pet', 0.6891531348228455),\n", | |||||
| " ('felines', 0.6755931377410889),\n", | |||||
| " ('chihuahua', 0.6709762215614319)]" | |||||
| ] | |||||
| }, | |||||
| "execution_count": 9, | |||||
| "metadata": {}, | |||||
| "output_type": "execute_result" | |||||
| } | |||||
| ], | |||||
| "source": [ | |||||
| "model.most_similar(\"cat\")" | |||||
| ] | |||||
| }, | |||||
| { | |||||
| "cell_type": "code", | |||||
| "execution_count": 16, | |||||
| "metadata": {}, | |||||
| "outputs": [ | |||||
| { | |||||
| "name": "stdout", | |||||
| "output_type": "stream", | |||||
| "text": [ | |||||
| "hi globe \n" | |||||
| ] | |||||
| } | |||||
| ], | |||||
| "source": [ | |||||
| "def transformSentence(sentence):\n", | |||||
| " outputSentence = \"\"\n", | |||||
| " \n", | |||||
| " for word in sentence.split(\" \"):\n", | |||||
| " try:\n", | |||||
| " outputSentence += model.most_similar(word)[0][0] + \" \"\n", | |||||
| " except Exception:\n", | |||||
| " outputSentence += word + \" \"\n", | |||||
| " return outputSentence\n", | |||||
| "\n", | |||||
| "print(transformSentence(\"hello world\"))" | |||||
| ] | |||||
| }, | |||||
| { | |||||
| "cell_type": "code", | |||||
| "execution_count": 12, | |||||
| "metadata": {}, | |||||
| "outputs": [ | |||||
| { | |||||
| "name": "stdout", | |||||
| "output_type": "stream", | |||||
| "text": [ | |||||
| "looks Mom No hand \n" | |||||
| ] | |||||
| } | |||||
| ], | |||||
| "source": [ | |||||
| "print(transformSentence(\"look mom no hands\"))" | |||||
| ] | |||||
| }, | |||||
| { | |||||
| "cell_type": "code", | |||||
| "execution_count": 17, | |||||
| "metadata": {}, | |||||
| "outputs": [ | |||||
| { | |||||
| "name": "stdout", | |||||
| "output_type": "stream", | |||||
| "text": [ | |||||
| "This gen_eral concept of Clustering was to groups Data wtih similiar trait \n" | |||||
| ] | |||||
| } | |||||
| ], | |||||
| "source": [ | |||||
| "print(transformSentence(\"The general idea of clustering is to group data with similar traits\"))" | |||||
| ] | |||||
| }, | |||||
| { | |||||
| "cell_type": "code", | |||||
| "execution_count": 52, | |||||
| "metadata": {}, | |||||
| "outputs": [ | |||||
| { | |||||
| "name": "stdout", | |||||
| "output_type": "stream", | |||||
| "text": [ | |||||
| "This manager concept of clusters was to groups datasets wtih similiar traits. \n" | |||||
| ] | |||||
| } | |||||
| ], | |||||
| "source": [ | |||||
| "def removeFromString(string, chars):\n", | |||||
| " for c in chars:\n", | |||||
| " string = string.replace(c, \"\")\n", | |||||
| " return string\n", | |||||
| "\n", | |||||
| "\n", | |||||
| "def transformSentenceWithHeuristic(sentence):\n", | |||||
| " outputSentence = \"\"\n", | |||||
| " \n", | |||||
| " for word in sentence.split(\" \"):\n", | |||||
| " try:\n", | |||||
| " changed = False\n", | |||||
| " for w, _ in model.most_similar(word):\n", | |||||
| " clean = removeFromString(w, [' ', '_']).lower()\n", | |||||
| " if clean not in word.lower() and \"_\" not in w:\n", | |||||
| " outputSentence += w + \" \"\n", | |||||
| " changed = True\n", | |||||
| " break\n", | |||||
| " outputSentence = outputSentence if changed else outputSentence + word + \" \"\n", | |||||
| " except Exception:\n", | |||||
| " outputSentence += word + \" \"\n", | |||||
| " return outputSentence\n", | |||||
| "print(transformSentenceWithHeuristic(\"The general idea of clustering is to group data with similar traits.\"))" | |||||
| ] | |||||
| }, | |||||
| { | |||||
| "cell_type": "code", | |||||
| "execution_count": 53, | |||||
| "metadata": {}, | |||||
| "outputs": [ | |||||
| { | |||||
| "name": "stdout", | |||||
| "output_type": "stream", | |||||
| "text": [ | |||||
| "Relax up and grabbing a drinks but that was day it I talking abut this hallucinogenic trips it was this 1981 film Fever Treatment. \n" | |||||
| ] | |||||
| } | |||||
| ], | |||||
| "source": [ | |||||
| "print(transformSentenceWithHeuristic(\"Sit down and grab a drink because it is time that we talk about the LSD trip that is the 1981 movie Shock Treatment.\"))" | |||||
| ] | |||||
| }, | |||||
| { | |||||
| "cell_type": "code", | |||||
| "execution_count": 54, | |||||
| "metadata": {}, | |||||
| "outputs": [], | |||||
| "source": [ | |||||
| "from sklearn.decomposition import IncrementalPCA # inital reduction\n", | |||||
| "from sklearn.manifold import TSNE # final reduction\n", | |||||
| "import numpy as np # array handling\n", | |||||
| "\n", | |||||
| "\n", | |||||
| "def reduce_dimensions(model):\n", | |||||
| " num_dimensions = 2 # final num dimensions (2D, 3D, etc)\n", | |||||
| "\n", | |||||
| " vectors = [] # positions in vector space\n", | |||||
| " labels = [] # keep track of words to label our data again later\n", | |||||
| " for word in model.wv.vocab:\n", | |||||
| " vectors.append(model.wv[word])\n", | |||||
| " labels.append(word)\n", | |||||
| "\n", | |||||
| " # convert both lists into numpy vectors for reduction\n", | |||||
| " vectors = np.asarray(vectors)\n", | |||||
| " labels = np.asarray(labels)\n", | |||||
| "\n", | |||||
| " # reduce using t-SNE\n", | |||||
| " vectors = np.asarray(vectors)\n", | |||||
| " tsne = TSNE(n_components=num_dimensions, random_state=0)\n", | |||||
| " vectors = tsne.fit_transform(vectors)\n", | |||||
| "\n", | |||||
| " x_vals = [v[0] for v in vectors]\n", | |||||
| " y_vals = [v[1] for v in vectors]\n", | |||||
| " return x_vals, y_vals, labels\n", | |||||
| "\n", | |||||
| "\n", | |||||
| "#x_vals, y_vals, labels = reduce_dimensions(model)" | |||||
| ] | |||||
| } | |||||
| ], | |||||
| "metadata": { | |||||
| "kernelspec": { | |||||
| "display_name": "Python 3", | |||||
| "language": "python", | |||||
| "name": "python3" | |||||
| }, | |||||
| "language_info": { | |||||
| "codemirror_mode": { | |||||
| "name": "ipython", | |||||
| "version": 3 | |||||
| }, | |||||
| "file_extension": ".py", | |||||
| "mimetype": "text/x-python", | |||||
| "name": "python", | |||||
| "nbconvert_exporter": "python", | |||||
| "pygments_lexer": "ipython3", | |||||
| "version": "3.8.1" | |||||
| } | |||||
| }, | |||||
| "nbformat": 4, | |||||
| "nbformat_minor": 4 | |||||
| } | |||||
+ 678
- 0
notebooks/word-embeddings/embedding-part-1.ipynb
File diff suppressed because it is too large
View File
BIN
notebooks/word-embeddings/swap.png
View File
+ 82
- 0
other/Dijkstra.py
View File
| @ -0,0 +1,82 @@ | |||||
| # Python program for Dijkstra's single | |||||
| # source shortest path algorithm. The program is | |||||
| # for adjacency matrix representation of the graph | |||||
| # Library for INT_MAX | |||||
| import sys | |||||
| class Graph(): | |||||
| def __init__(self, vertices): | |||||
| self.V = vertices | |||||
| self.graph = [[0 for column in range(vertices)] | |||||
| for row in range(vertices)] | |||||
| def printSolution(self, dist): | |||||
| print "Vertex tDistance from Source" | |||||
| for node in range(self.V): | |||||
| print node, "t", dist[node] | |||||
| # A utility function to find the vertex with | |||||
| # minimum distance value, from the set of vertices | |||||
| # not yet included in shortest path tree | |||||
| def minDistance(self, dist, sptSet): | |||||
| # Initilaize minimum distance for next node | |||||
| min = sys.maxint | |||||
| # Search not nearest vertex not in the | |||||
| # shortest path tree | |||||
| for v in range(self.V): | |||||
| if dist[v] < min and sptSet[v] == False: | |||||
| min = dist[v] | |||||
| min_index = v | |||||
| return min_index | |||||
| # Funtion that implements Dijkstra's single source | |||||
| # shortest path algorithm for a graph represented | |||||
| # using adjacency matrix representation | |||||
| def dijkstra(self, src): | |||||
| dist = [sys.maxint] * self.V | |||||
| dist[src] = 0 | |||||
| sptSet = [False] * self.V | |||||
| for cout in range(self.V): | |||||
| # Pick the minimum distance vertex from | |||||
| # the set of vertices not yet processed. | |||||
| # u is always equal to src in first iteration | |||||
| u = self.minDistance(dist, sptSet) | |||||
| # Put the minimum distance vertex in the | |||||
| # shotest path tree | |||||
| sptSet[u] = True | |||||
| # Update dist value of the adjacent vertices | |||||
| # of the picked vertex only if the current | |||||
| # distance is greater than new distance and | |||||
| # the vertex in not in the shotest path tree | |||||
| for v in range(self.V): | |||||
| if self.graph[u][v] > 0 and sptSet[v] == False and \ | |||||
| dist[v] > dist[u] + self.graph[u][v]: | |||||
| dist[v] = dist[u] + self.graph[u][v] | |||||
| self.printSolution(dist) | |||||
| # Driver program | |||||
| g = Graph(9) | |||||
| g.graph = [[0, 4, 0, 0, 0, 0, 0, 8, 0], | |||||
| [4, 0, 8, 0, 0, 0, 0, 11, 0], | |||||
| [0, 8, 0, 7, 0, 4, 0, 0, 2], | |||||
| [0, 0, 7, 0, 9, 14, 0, 0, 0], | |||||
| [0, 0, 0, 9, 0, 10, 0, 0, 0], | |||||
| [0, 0, 4, 14, 10, 0, 2, 0, 0], | |||||
| [0, 0, 0, 0, 0, 2, 0, 1, 6], | |||||
| [8, 11, 0, 0, 0, 0, 1, 0, 7], | |||||
| [0, 0, 2, 0, 0, 0, 6, 7, 0] | |||||
| ]; | |||||
| g.dijkstra(0); | |||||
+ 52
- 0
other/knapSack.py
View File
| @ -0,0 +1,52 @@ | |||||
| """ | |||||
| Jeffery Russell | |||||
| 12-1-18 | |||||
| """ | |||||
| def knapsack(V, W, capacity): | |||||
| """ | |||||
| Dynamic programming implementation of the knapsack problem | |||||
| :param V: List of the values | |||||
| :param W: List of weights | |||||
| :param capacity: max capacity of knapsack | |||||
| :return: List of tuples of objects stolen in form (w, v) | |||||
| """ | |||||
| choices = [[[] for i in range(capacity + 1)] for j in range(len(V) + 1)] | |||||
| cost = [[0 for i in range(capacity + 1)] for j in range(len(V) + 1)] | |||||
| for i in range(0, len(V)): | |||||
| for j in range(0, capacity + 1): | |||||
| if W[i] > j: # don't include another item | |||||
| cost[i][j] = cost[i -1][j] | |||||
| choices[i][j] = choices[i - 1][j] | |||||
| else: # Adding another item | |||||
| cost[i][j] = max(cost[i-1][j], cost[i-1][j - W[i]] + V[i]) | |||||
| if cost[i][j] != cost[i-1][j]: | |||||
| choices[i][j] = choices[i - 1][j - W[i]] + [(W[i], V[i])] | |||||
| else: | |||||
| choices[i][j] = choices[i - 1][j] | |||||
| return choices[len(V) -1][capacity] | |||||
| def printSolution(S): | |||||
| """ | |||||
| Takes the output of knapsack and prints it in a | |||||
| pretty format. | |||||
| :param S: list of tuples representing items stolen | |||||
| :return: None | |||||
| """ | |||||
| print("Thief Took:") | |||||
| for i in S: | |||||
| print("Weight: " + str(i[0]) + "\tValue: \t" + str(i[1])) | |||||
| print() | |||||
| print("Total Value Stolen: " + str(sum(int(v[0]) for v in S))) | |||||
| print("Total Weight in knapsack: " + str(sum(int(v[1]) for v in S))) | |||||
| values = [99,1,1,1,1, 1,1] | |||||
| weights = [5,1,2,3,4,5,1] | |||||
| printSolution(knapsack(values, weights, 6)) | |||||
+ 129
- 0
other/markdownParagraphFormatter.py
View File
| @ -0,0 +1,129 @@ | |||||
| """ | |||||
| Jeffery Russell | |||||
| 2019-4-21 | |||||
| Simple program to reformat paragraphs in markdown text files | |||||
| to follow a specific col limit. | |||||
| """ | |||||
| import os | |||||
| import sys | |||||
| def formatChunk(stringContent, charLimit): | |||||
| """ | |||||
| Formats a markdown section with with a specific | |||||
| col limit. This only makes changes if the section is a | |||||
| paragraph and nothing else. | |||||
| The only guarantee for the input is that there | |||||
| are no lines which only contain a new line or space. | |||||
| :param stringContent: string of markdown chunk | |||||
| :param charLimit: max character limit | |||||
| :return: formatted string | |||||
| """ | |||||
| if len(stringContent.strip(" ")) == 0: | |||||
| return "" | |||||
| elif not stringContent[0].isalpha(): | |||||
| return stringContent | |||||
| result = "" | |||||
| line = "" | |||||
| stringContent = stringContent.replace("\n", " ") | |||||
| words = stringContent.split(" ") | |||||
| for word in words: | |||||
| if len(line) == 0 or len(line) + len(word) < charLimit: | |||||
| if not len(line) == 0: | |||||
| line = line + " " | |||||
| line = line + word | |||||
| else: | |||||
| result = result + line + "\n" | |||||
| line = word | |||||
| if line != "": | |||||
| result = result + line + "\n" | |||||
| return result | |||||
| def writeToFile(fileName, content): | |||||
| """ | |||||
| Writes to a file, overwriting the existing | |||||
| output. | |||||
| :param fileName: | |||||
| :param content: | |||||
| :return: | |||||
| """ | |||||
| f = open(fileName, "w+") | |||||
| f.write(content) | |||||
| f.close() | |||||
| def wrapMarkdownParagraphsToColLimit(fileName, charLimit): | |||||
| """ | |||||
| Breaks apart a markdown file into chunks. A chunks are | |||||
| separated by a blank lines. Each chunk is then | |||||
| rendered according to our 80 char rules defined | |||||
| in the formatChunk section. | |||||
| :param fileName: | |||||
| :param charLimit: | |||||
| :return: | |||||
| """ | |||||
| result = "" | |||||
| tempResult = "" | |||||
| inCodeBlock = False | |||||
| with open(fileName) as file: | |||||
| for line in file: | |||||
| if line.startswith("```"): | |||||
| inCodeBlock = not inCodeBlock | |||||
| result = result + line | |||||
| elif inCodeBlock: | |||||
| result = result + line | |||||
| elif line.strip(" ") == "\n": | |||||
| result = result + formatChunk(tempResult, charLimit) | |||||
| tempResult = "" | |||||
| result = result + "\n" | |||||
| else: | |||||
| tempResult = tempResult + line | |||||
| if tempResult != "": | |||||
| result = result + formatChunk(tempResult, charLimit) | |||||
| return result | |||||
| def print_usage(): | |||||
| """ | |||||
| Prints script usage. | |||||
| :return: | |||||
| """ | |||||
| print("Usage: file name -t (optional)") | |||||
| print("\t-p simply prints the formatted output.") | |||||
| print("\tfile name -- runs the command overwriting the existing file.") | |||||
| def main(): | |||||
| """ | |||||
| Checks command line input and calls the proper formatting | |||||
| functions. | |||||
| :return: | |||||
| """ | |||||
| if len(sys.argv) > 1: | |||||
| fileName = os.getcwd() + "/" + sys.argv[1] | |||||
| if len(sys.argv) == 3 and sys.argv[2].lower() == "-p": | |||||
| print(wrapMarkdownParagraphsToColLimit(fileName, 70)) | |||||
| else: | |||||
| writeToFile(fileName, | |||||
| wrapMarkdownParagraphsToColLimit(fileName, 70)) | |||||
| else: | |||||
| print_usage() | |||||
| """ | |||||
| Makes sure that other scripts don't execute the main | |||||
| """ | |||||
| if __name__ == '__main__': | |||||
| try: | |||||
| main() | |||||
| except KeyboardInterrupt: | |||||
| panic() | |||||
+ 14
- 3
other/minMul.py
View File
+ 162
- 0
other/paragraphFormatting.py
View File
| @ -0,0 +1,162 @@ | |||||
| import math | |||||
| """ | |||||
| Jeffery Russell | |||||
| 11-13-18 | |||||
| """ | |||||
| def extraSpace(S, M, i, j): | |||||
| """ | |||||
| Computes the number of extra characters at the end of | |||||
| the line. | |||||
| Between each word there is only once space. | |||||
| :param S: List of words | |||||
| :param M: Max length of line | |||||
| :param i: start word index | |||||
| :param j: end word index | |||||
| """ | |||||
| extraSpaces = M - j + i | |||||
| for x in range(i, j + 1): | |||||
| extraSpaces -= len(S[x]) | |||||
| return extraSpaces | |||||
| def badnessLine(S, M, i, j): | |||||
| """ | |||||
| Computes Line badness. This is the number of | |||||
| extra spaces or infinity if the length exceeds M | |||||
| :param S: List of words | |||||
| :param M: Max length of line | |||||
| :param i: start word index | |||||
| :param j: end word index | |||||
| """ | |||||
| es = extraSpace(S, M, i, j) | |||||
| if es < 0: | |||||
| return math.inf | |||||
| return es | |||||
| def minBad(S, M, i): | |||||
| """ | |||||
| Computes the badness of a paragraph as the | |||||
| badness of the worst line in the paragraph not | |||||
| including the last line. | |||||
| *this is recursive | |||||
| :param S: List of words | |||||
| :param M: Max length of line | |||||
| :param i: start word index | |||||
| """ | |||||
| if badnessLine(S, M, i, len(S) -1) != math.inf: | |||||
| return 0 | |||||
| min = math.inf | |||||
| for k in range(i + 1, len(S)): | |||||
| end = minBad(S, M, k) | |||||
| front = badnessLine(S, M, i, k - 1) | |||||
| max = end if end > front else front | |||||
| min = min if min < max else max | |||||
| return min | |||||
| def minBadDynamic(S, M): | |||||
| """ | |||||
| Write a procedure minBadDynamic that implements | |||||
| the function mb' using dynamic program- | |||||
| ming. It should take only two parameters: S and M | |||||
| :param S: List of words | |||||
| :param M: Max length of line | |||||
| """ | |||||
| cost = [math.inf for i in range(len(S))] | |||||
| for i in range(0, len(S)): | |||||
| if badnessLine(S, M, 0, i) != math.inf: | |||||
| cost[i] = badnessLine(S, M, 0, i) | |||||
| if i == len(S) -1: | |||||
| return 0 #Everything fit on one line | |||||
| else: | |||||
| min = math.inf | |||||
| for k in range(0, i): | |||||
| before = cost[k] | |||||
| after = badnessLine(S, M, k + 1, i) | |||||
| if i == len(S) -1 and badnessLine(S, M, k+1, i) != math.inf: | |||||
| after = 0 # Last Line | |||||
| max = before if before > after else after | |||||
| min = min if min < max else max | |||||
| cost[i] = min | |||||
| return cost[len(S) -1] | |||||
| def minBadDynamicChoice(S, M): | |||||
| """ | |||||
| Write a procedure minBadDynamicChoice that implements | |||||
| the function mb' using dynamic | |||||
| programming. In addition to returning mb(S, M ), it | |||||
| should also return the choices made | |||||
| :param S: List of words | |||||
| :param M: Max length of line | |||||
| """ | |||||
| cost = [math.inf for i in range(len(S))] | |||||
| # List of tuples indicating start/end indices of line | |||||
| choice = [[] for i in range(len(S))] | |||||
| for i in range(0, len(S)): | |||||
| if badnessLine(S, M, 0, i) != math.inf: | |||||
| cost[i] = badnessLine(S, M, 0, i) | |||||
| choice[i] = [(0, i)] | |||||
| if i == len(S) - 1: | |||||
| return 0, [(0,i)] # One line | |||||
| else: | |||||
| min = math.inf | |||||
| choiceCanidate = [] | |||||
| for k in range(0, i): # Finds the optimal solution | |||||
| before = cost[k] # Previously computed minimum | |||||
| after = badnessLine(S, M, k + 1, i) # Badness of new slice | |||||
| if i == len(S) - 1 and badnessLine(S, M, k+1, i) != math.inf: | |||||
| after = 0 # Last line | |||||
| max = before if before > after else after | |||||
| if min > max: | |||||
| # Captures where slice is being taken | |||||
| choiceCanidate = choice[k] + [(k+1, i)] | |||||
| min = max | |||||
| choice[i] = choiceCanidate | |||||
| cost[i] = min | |||||
| return cost[len(S) -1], choice[len(S) -1] | |||||
| def printParagraph(S, M): | |||||
| """ | |||||
| which takes two parameters: S and M that displays the words in S on | |||||
| the screen using the choices of minBadDynamicChoice. | |||||
| What is the asymptotic running time of your | |||||
| algorithm? | |||||
| This program will run asymptotically in O(n^2) due | |||||
| to the characteristic of calculating the minBad | |||||
| for each sub sequence and then looping through a | |||||
| maximum of |S| to find the optimal solution. | |||||
| :param S: List of words | |||||
| :param M: Max length of line | |||||
| """ | |||||
| cost, choice = minBadDynamicChoice(S, M) | |||||
| print() | |||||
| for i in range(0, len(choice)): | |||||
| for x in range(choice[i][0], choice[i][1] + 1): | |||||
| print(str(S[x]), end=" ") | |||||
| print() | |||||
| print(minBadDynamicChoice(["aaa", "aaa"], 6)) | |||||
| printParagraph(["jjjjjj","aaa", "bb", "cc", "ff", "mm", "ddddd", "ddddd"], 6) | |||||
| printParagraph(["jjjjjj"], 6) | |||||
+ 65
- 0
perceptron/perceptron.py
View File
| @ -0,0 +1,65 @@ | |||||
| import numpy as np | |||||
| class Perceptron(object): | |||||
| """ | |||||
| Perceptron classifier | |||||
| ___________________ | |||||
| parameters | |||||
| __________________ | |||||
| eta: float | |||||
| learning rate betweeen 0.0 and 1.0 | |||||
| n_iter: int | |||||
| Passes over the training dataset | |||||
| ___________________ | |||||
| Attributes | |||||
| ___________________ | |||||
| w_: 1d array | |||||
| weights after training | |||||
| errors_: list | |||||
| Number of msiclassifications for every epoch | |||||
| """ | |||||
| def __init__(self, eta, n_iter): | |||||
| self.eta = eta | |||||
| self.n_iter = n_iter | |||||
| def fit(self, X, y): | |||||
| """ | |||||
| Fit training data | |||||
| ______________________ | |||||
| parameters | |||||
| ______________________ | |||||
| X: {array-like}, shape = [n_samples, n_features] | |||||
| Training vectors where n_samples is the number of samples | |||||
| and n_features is the number of features | |||||
| y: array-like, shape = [n_samples] | |||||
| Target values | |||||
| ____________________ | |||||
| Returns | |||||
| ____________________ | |||||
| self: object | |||||
| """ | |||||
| self.w_ = np.zeros(1 + X.shape[1]) | |||||
| self.errors_ = [] | |||||
| for _ in range(self.n_iter): | |||||
| errors = 0 | |||||
| for xi,target in zip(X, y): | |||||
| update = self.eta * (target - self.predict(xi)) | |||||
| self.w_[1:] += update * xi | |||||
| self.w_[0] += update | |||||
| errors += int(update != 0.0) | |||||
| self.errors_.append(errors) | |||||
| return self | |||||
| def net_input(self, X): | |||||
| """ | |||||
| Calculate net input | |||||
| """ | |||||
| return np.dot(X, self.w_[1:]) + self.w_[0] | |||||
| def predict(self, X): | |||||
| """ | |||||
| Return class label after unit step | |||||
| """ | |||||
| return np.where(self.net_input(X) >= 0.0, 1, -1) | |||||
+ 42
- 0
searching_algo/BFS.py
View File
| @ -0,0 +1,42 @@ | |||||
| from collections import defaultdict | |||||
| #--class for directed graph with adjacency list representation | |||||
| class Graph: | |||||
| #--Constructor of the class | |||||
| def __init__(self): | |||||
| #--default dictionary to store graph | |||||
| self.graph = defaultdict(list) | |||||
| ''' | |||||
| --function to add an edge | |||||
| --inputs, starting point of the graph | |||||
| ''' | |||||
| def addEdge(self,u,v): | |||||
| self.graph[u].append(v) | |||||
| ''' | |||||
| --to print a BFS of graph | |||||
| --inputs, starting point of the graph | |||||
| --Prints the BFS starting from the input point | |||||
| ''' | |||||
| def BFS(self, s): | |||||
| # Mark all the vertices as not visited | |||||
| visited = [False] * (len(self.graph)) | |||||
| queue = [] | |||||
| # Mark the source node as visited and enqueue it | |||||
| queue.append(s) | |||||
| visited[s] = True | |||||
| while queue: | |||||
| s = queue.pop(0) | |||||
| print (s, end = " ") | |||||
| for i in self.graph[s]: | |||||
| if visited[i] == False: | |||||
| queue.append(i) | |||||
| visited[i] = True | |||||
+ 39
- 0
searching_algo/Jump_Search.py
View File
| @ -0,0 +1,39 @@ | |||||
| import math | |||||
| def jumpSearch( arr , x ): | |||||
| # Finding block size to be jumped | |||||
| n = len(arr) | |||||
| step = math.sqrt(n) | |||||
| # Finding the block where element is | |||||
| # present (if it is present) | |||||
| prev = 0 | |||||
| while arr[int(min(step, n)-1)] < x: | |||||
| prev = step | |||||
| step += math.sqrt(n) | |||||
| if prev >= n: | |||||
| return -1 | |||||
| # Doing a linear search for x in | |||||
| # block beginning with prev. | |||||
| while arr[int(prev)] < x: | |||||
| prev += 1 | |||||
| # If we reached next block or end | |||||
| # of array, element is not present. | |||||
| if prev == min(step, n): | |||||
| return -1 | |||||
| # If element is found | |||||
| if arr[int(prev)] == x: | |||||
| return int(prev) | |||||
| return -1 | |||||
| arr = [ 0, 1, 3, 4, 4, 5, 8, 13, 22, 24, 55, 59, 122, 213, 422, 555 ] | |||||
| x = 55 | |||||
| index = jumpSearch(arr, x) | |||||
| print("Number is at index {}".format(index)) | |||||
+ 33
- 0
searching_algo/exponentialSearch.py
View File
| @ -0,0 +1,33 @@ | |||||
| def search(arr, l, r, x): | |||||
| if r >= l: | |||||
| m = (l+r)/2 | |||||
| if arr[m] == x: | |||||
| return m | |||||
| if arr[m] > x: | |||||
| return search(arr, l, m-1, x) | |||||
| else: | |||||
| return search(arr, m, r, x) | |||||
| return -1 | |||||
| def exponentialSearch(arr, n, x): | |||||
| if arr[0] == x: | |||||
| return 0 | |||||
| i = 1 | |||||
| while i<n and arr[i] <=x: | |||||
| i = i*2 | |||||
| return search(arr, i/2, min(n, i), x) | |||||
| arr = [2, 3, 4, 7, 10, 40] | |||||
| n = len(arr) | |||||
| x = 10 | |||||
| r = exponentialSearch(arr, n, x) | |||||
| if r == -1: | |||||
| print "Element not found" | |||||
| else: | |||||
| print "Element is present at index %d" %(r) | |||||
+ 35
- 0
sorting/bucketSort.py
View File
| @ -0,0 +1,35 @@ | |||||
| def bucket_sort(alist): | |||||
| largest = max(alist) | |||||
| length = len(alist) | |||||
| size = largest/length | |||||
| buckets = [[] for _ in range(length)] | |||||
| for i in range(length): | |||||
| j = int(alist[i]/size) | |||||
| if j != length: | |||||
| buckets[j].append(alist[i]) | |||||
| else: | |||||
| buckets[length - 1].append(alist[i]) | |||||
| for i in range(length): | |||||
| insertion_sort(buckets[i]) | |||||
| result = [] | |||||
| for i in range(length): | |||||
| result = result + buckets[i] | |||||
| return result | |||||
| def insertion_sort(alist): | |||||
| for i in range(1, len(alist)): | |||||
| temp = alist[i] | |||||
| j = i - 1 | |||||
| while (j >= 0 and temp < alist[j]): | |||||
| alist[j + 1] = alist[j] | |||||
| j = j - 1 | |||||
| alist[j + 1] = temp | |||||
| alist = [54,26,93,17,77,31,44,55,20] | |||||
| print(bucket_sort(alist)) | |||||
+ 122
- 0
sorting/heapsort.py
View File
| @ -0,0 +1,122 @@ | |||||
| """ | |||||
| :Author: James Sherratt | |||||
| :Date: 20/10/2019 | |||||
| :License: MIT | |||||
| :name: heapsort.py | |||||
| Heap sorts a list-like object. Note: this has been written with code-clarity | |||||
| in mind first, efficiency second. | |||||
| """ | |||||
| from random import randint | |||||
| def get_left(i): | |||||
| """ | |||||
| Get the left element index of a heap node for an array. | |||||
| :param i: The parent index. | |||||
| :return: the left element. | |||||
| """ | |||||
| return 2 * i + 1 | |||||
| def get_right(i): | |||||
| """ | |||||
| Get the right element index of a heap node for an array. | |||||
| :param i: The parent index. | |||||
| :return: the right element. | |||||
| """ | |||||
| return 2 * i + 2 | |||||
| def repair_heap(vals_list, root, arr_top): | |||||
| """ | |||||
| Sifts the root element of a heap to the correct position, to | |||||
| correct a max heap. This assumes the children of the root/ node are max heaps. | |||||
| :param vals_list: list of values, which represents a heap structure. | |||||
| :param root: the index of the node we're working from/ using as a root. | |||||
| :param arr_top: the largest value of the list we're interested in. | |||||
| :return: Reference to the passed list, with the root node in the correct position. | |||||
| """ | |||||
| # This is the value to swap. We want to swap the root value down, so we swap the root first. | |||||
| swap = root | |||||
| # Get left and right nodes of root. | |||||
| left = get_left(root) | |||||
| right = get_right(root) | |||||
| while left < arr_top: | |||||
| # Check if value to swap is less than the left child. | |||||
| if vals_list[swap] < vals_list[left]: | |||||
| swap = left | |||||
| # Check if value to swap is less than the right child (if exists). | |||||
| # Note: these 2 if's could be combined using "and", but then we're relying on lazy evaluation. | |||||
| if right < arr_top: | |||||
| if vals_list[swap] < vals_list[right]: | |||||
| swap = right | |||||
| # Check if the swap is still the root. If so, there's no more children to swap and we're done. | |||||
| if swap == root: | |||||
| return vals_list | |||||
| # Else, swap. | |||||
| else: | |||||
| vals_list[root], vals_list[swap] = vals_list[swap], vals_list[root] | |||||
| # New root, left and right node for the next iteration. | |||||
| root = swap | |||||
| left = get_left(root) | |||||
| right = get_right(root) | |||||
| return vals_list | |||||
| def max_heap(vals_list): | |||||
| """ | |||||
| Convert a list of values into a max heap tree. | |||||
| :param vals_list: list of numbers. | |||||
| :return: the same list as a max heap tree. | |||||
| """ | |||||
| # Create a max heap by repairing the heap, starting from the nodes one above the leaf nodes. | |||||
| len_list = len(vals_list) | |||||
| for root in range(len_list//2, -1, -1): | |||||
| repair_heap(vals_list, root, len_list) | |||||
| return vals_list | |||||
| def max_heap_to_sorted(vals_list): | |||||
| """ | |||||
| Convert a max heap list into a sorted list. | |||||
| :param vals_list: list containing max heap. | |||||
| :return: the same list of values, sorted. | |||||
| """ | |||||
| # i is the index of the last element of the slice of the array that needs sorting. | |||||
| for top in range(len(vals_list)-1, 0, -1): | |||||
| # Swap the root value (max) with the last value of the slice. | |||||
| vals_list[0], vals_list[top] = vals_list[top], vals_list[0] | |||||
| # Sift the new root to the correct position of the remainder of the max heap. | |||||
| # Another way of doing this is to pass a slice of the vals_list up to the value top, but python passes | |||||
| # slices by copy so there's a massive performance hit. | |||||
| repair_heap(vals_list, 0, top) | |||||
| return vals_list | |||||
| def heapsort(vals_list): | |||||
| """ | |||||
| Sort a list of values using heapsort. | |||||
| :param vals_list: list of sortable values. | |||||
| :return: the same list, sorted. | |||||
| """ | |||||
| max_heap(vals_list) | |||||
| return max_heap_to_sorted(vals_list) | |||||
| if __name__ == "__main__": | |||||
| list_len = 100000 | |||||
| vals_list = [randint(0, (2**16)) for i in range(list_len)] | |||||
| heap_sorted = heapsort(list(vals_list)) | |||||
| py_sorted = sorted(vals_list) | |||||
| print("Did the sort work? {}".format(heap_sorted == py_sorted)) | |||||
+ 39
- 0
text_preprocessing.py
View File
| @ -0,0 +1,39 @@ | |||||
| import clean_text | |||||
| # import all our functions | |||||
| from clean_text import * | |||||
| #!pylint cleantext | |||||
| import pandas as pd | |||||
| from sklearn.feature_extraction.text import CountVectorizer | |||||
| training = [ | |||||
| " I am master of all", | |||||
| "I am a absolute learner" | |||||
| ] | |||||
| generalization = [ | |||||
| "I am absolute learner learner" | |||||
| ] | |||||
| vectorization = CountVectorizer( | |||||
| stop_words = "english", | |||||
| preprocessor = process.master_clean_text) | |||||
| vectorization.fit(training) | |||||
| build_vocab = { | |||||
| value:key | |||||
| for key , value in vectorization.vocabulary_.items() | |||||
| } | |||||
| vocab = [build_vocab[i] for i in range(len(build_vocab))] | |||||
| extracted = pd.DataFrame( | |||||
| data = vectorization.transform(generalization).toarray(), | |||||
| index=["generalization"], | |||||
| columns=vocab | |||||
| ) | |||||
| print(extracted) | |||||
+ 14
- 0
windows/Sticky Notes Backuper/Readme.txt
View File
| @ -0,0 +1,14 @@ | |||||
| This is a very simple program. | |||||
| Double click on grab StickyNotes.bat to backup your sticky notes | |||||
| into the current directory. | |||||
| Double click ReplaceCurrentStickyNotes.bat to replace your | |||||
| stickynotes with the ones in the current directory. | |||||
| ** Note this will only work with windows 10 and up. | |||||
| Windows 10 uses the .sqlite format where windows 7 | |||||
| used a .snt file. | |||||
+ 9
- 0
windows/Sticky Notes Backuper/grabStickyNotes.bat
View File
| @ -0,0 +1,9 @@ | |||||
| :: Author : Jeffery Russell 11-27-18 | |||||
| @echo off | |||||
| pushd %~dp0 | |||||
| dir | |||||
| copy %LocalAppData%\Packages\Microsoft.MicrosoftStickyNotes_8wekyb3d8bbwe\LocalState\plum.sqlite | |||||
| pause | |||||
+ 9
- 0
windows/Sticky Notes Backuper/replaceCurrentStickyNotes.bat
View File
| @ -0,0 +1,9 @@ | |||||
| :: Author : Jeffery Russell 11-27-18 | |||||
| @echo off | |||||
| pushd %~dp0 | |||||
| dir | |||||
| copy plum.sqlite %LocalAppData%\Packages\Microsoft.MicrosoftStickyNotes_8wekyb3d8bbwe\LocalState\plum.sqlite | |||||
| pause | |||||