5 changed files with 212 additions and 0 deletions
Unified View
Diff Options
-
BINblogContent/headerImages/steamGraphs2.png
-
BINblogContent/posts/data-science/media/steamNode2vec/friends.png
-
BINblogContent/posts/data-science/media/steamNode2vec/games.png
-
BINblogContent/posts/data-science/media/steamNode2vec/output_9_0.png
-
+212 -0blogContent/posts/data-science/node2vec-with-steam-data.md
BIN
blogContent/headerImages/steamGraphs2.png
View File
BIN
blogContent/posts/data-science/media/steamNode2vec/friends.png
View File
BIN
blogContent/posts/data-science/media/steamNode2vec/games.png
View File
BIN
blogContent/posts/data-science/media/steamNode2vec/output_9_0.png
View File
+ 212
- 0
blogContent/posts/data-science/node2vec-with-steam-data.md
View File
| @ -0,0 +1,212 @@ | |||||
| Graph algorithms!!! | |||||
| Working with graphs can be a great deal of fun, but, sometimes we just want some cold hard vectors to do some good old fashion machine learning. | |||||
| This post looks at the famous [node2vec](https://cs.stanford.edu/~jure/pubs/node2vec-kdd16.pdf) algorithm used to quantize graph data. | |||||
| The example I'm giving in this blog post uses data from my recently resurrected [steam graph project](https://jrtechs.net/projects/steam-friends-graph). | |||||
| If you live under a rock, [Steam](https://store.steampowered.com/) is a platform where users can purchase, manage, and play games with friends. | |||||
| Although there is a ton of data within the Steam network, I am only interested in the graphs formed connecting users, friends, and games. | |||||
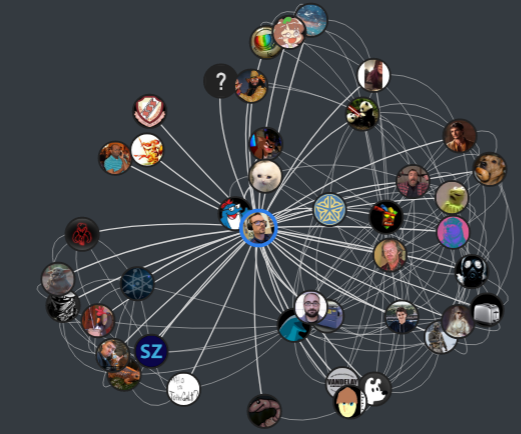
| My updated visualization to show a friendship network looks like this: | |||||
|  | |||||
| I'm also working on visualizations to show both friends and their games. | |||||
|  | |||||
| The issue that I'm currently running into is that these graphs quickly become egregiously large. Players on Steam frequently have 100+ friends and own over 50 games-- one person I indexed somehow had over 400 games! | |||||
| The size of this graph balloons exponentially with traversal depth. | |||||
| The sheer scale of the graph data makes it challenging to visualize concisely. | |||||
| Visualization of graph data brings us to our topic today: | |||||
| # Node2Vec | |||||
| Node2Vec is an embedding algorithm inspired by [word2vec](https://jrtechs.net/data-science/word-embeddings). | |||||
| The goal of this algorithm is to covert every node in a graph into a vectorized output where points close in the latent space correspond to related nodes. | |||||
| Simplifying a few things: node2vec uses the notion of a biased random walk through the graph. | |||||
| Two parameters -P and Q- defines whether we want to favor a BFS vs. a DFS traversal. A BFS search will give us a better local view of the network where a DFS traversal will provide us with a more global view of the graph. Applying some maths to the random walker outputs, distance in this embedding space is correlated to the probability that two nodes co-occur on the same random walk over the network. | |||||
| If you have the time, I urge you to watch [Jure Leskovec's](https://scholar.google.com/citations?user=Q_kKkIUAAAAJ&hl=en) lectures on graph learning on Youtube: | |||||
| <youtube src="YrhBZUtgG4E" /> | |||||
| Additionally, if you want to dive deeper into graph learning, I suggest that you dig through the Stanford [CS-224 course page](https://github.com/jrtechs/cs224w-notes). | |||||
| # Python Node2Vec Code | |||||
| I'm using a simple implementation of Node2Vec that I found on GitHub: [aditya-grover/node2vec](https://github.com/aditya-grover/node2vec). | |||||
| I'm using this package because it is a faithful implementation to the original paper and doesn't require you to install a lot of dependencies. This was written in Python 2, so to get Python 3 support, you will need to merge in changes from someone's fork because the maintainer is not reviewing any of the pull requests. | |||||
| ``` | |||||
| git clone https://github.com/aditya-grover/node2vec | |||||
| git remote add python3 https://github.com/mcwehner/node2vec | |||||
| git pull python3 master | |||||
| git pull python3 updated_requirements | |||||
| ``` | |||||
| Pip makes installing the dependencies easy using a requirements file. | |||||
| ```python | |||||
| !pip install -r node2vec/requirements.txt | |||||
| ``` | |||||
| Before we run this algorithm, we need to generate some data. | |||||
| An example edge list is in the [git repo](https://github.com/aditya-grover/node2vec/blob/master/graph/karate.edgelist). | |||||
| Using the JanusGraph database I'm using for the [Steam graphs project](https://github.com/jrtechs/SteamFriendsGraph), I generated an edge list for a single player's network. | |||||
| The code is straightforward; first, I pull the steam ids of the people that I want in my embedding graph. | |||||
| Second, I pull all the friend connections for the people that I want in the graph, and I filter out any players that are not already in the network. | |||||
| Finally, I take all the friend relationships and generate the edge pairs and save it to a file. If you are not familiar with data streaming in Java, I suggest that you check out my [last blog post](https://jrtechs.net/java/fun-with-functional-java). | |||||
| ```java | |||||
| Set<String> importantEdges = graph | |||||
| .getPlayer(baseID) | |||||
| .getFriends() | |||||
| .parallelStream() | |||||
| .map(Player::getId) | |||||
| .collect(Collectors.toSet()); | |||||
| Map<String, Set<String>> edgeList = new HashMap<String, Set<String>>() | |||||
| {{ | |||||
| importantEdges.forEach(f -> | |||||
| put(f, | |||||
| graph.getPlayer(f) | |||||
| .getFriends() | |||||
| .parallelStream() | |||||
| .map(Player::getId) | |||||
| .filter(importantEdges::contains) | |||||
| .collect(Collectors.toSet())) | |||||
| ); | |||||
| }}; | |||||
| List<String> edges = edgeList.keySet() | |||||
| .parallelStream().map(k -> | |||||
| edgeList.get(k) | |||||
| .stream() | |||||
| .map(k2 -> k + " " + k2) | |||||
| .collect(Collectors.toList())) | |||||
| .flatMap(Collection::stream) | |||||
| .collect(Collectors.toList()); | |||||
| WrappedFileWriter.writeToFileFromList(edges, outFile); | |||||
| ``` | |||||
| Using the edge list generated in the prior script, I feed it into the node2vec program. | |||||
| ```python | |||||
| !python node2vec/src/main.py --input jrtechs.edgelist --output output/jrtechs2.emd --num-walks=40 --dimensions=50 | |||||
| output: | |||||
| Walk iteration: | |||||
| 1 / 40 | |||||
| ... | |||||
| 40 / 40 | |||||
| ``` | |||||
| Once we have our embedding file, we can load it into Python to do machine learning or visualization. | |||||
| The output of the node2vec algorithm is a sequence of lines where each line starts with the node label, and the rest of the line is the embedding vector. | |||||
| ```python | |||||
| labels=[] | |||||
| vectors=[] | |||||
| with open("output/jrtechs2.emd") as fp: | |||||
| for line in fp: | |||||
| l_list = list(map(float, line.split())) | |||||
| vectors.append(l_list[1::]) | |||||
| labels.append(line.split()[0]) | |||||
| ``` | |||||
| Right now, I am interested in visualizing the output. However, that is impractical since it has 50 dimensions! Using the TSNE method, we can reduce the dimensionality so that we can visualize it. | |||||
| ```python | |||||
| from sklearn.decomposition import IncrementalPCA # inital reduction | |||||
| from sklearn.manifold import TSNE # final reduction | |||||
| import numpy as np | |||||
| def reduce_dimensions(labels, vectors, num_dimensions=2): | |||||
| # convert both lists into numpy vectors for reduction | |||||
| vectors = np.asarray(vectors) | |||||
| labels = np.asarray(labels) | |||||
| # reduce using t-SNE | |||||
| vectors = np.asarray(vectors) | |||||
| tsne = TSNE(n_components=num_dimensions, random_state=0) | |||||
| vectors = tsne.fit_transform(vectors) | |||||
| x_vals = [v[0] for v in vectors] | |||||
| y_vals = [v[1] for v in vectors] | |||||
| return x_vals, y_vals, labels | |||||
| x_vals, y_vals, labels = reduce_dimensions(labels, vectors) | |||||
| ``` | |||||
| Before we visualize our data, we will want to grab the name of the steam users because right now, all we have is their steam id, which is just a unique number. | |||||
| Back in our JanusGraph, we can quickly export the steam ids mapped to the players' names. | |||||
| ```java | |||||
| Player player = graph.getPlayer(id); | |||||
| List<String> names = new ArrayList<String>() | |||||
| {{ | |||||
| add(id + " " + player.getName()); | |||||
| addAll( | |||||
| player.getFriends() | |||||
| .stream() | |||||
| .map(p -> p.getId() + " " + p.getName()) | |||||
| .collect(Collectors.toList()) | |||||
| ); | |||||
| }}; | |||||
| WrappedFileWriter.writeToFileFromList(names, "friendsMap.map"); | |||||
| ``` | |||||
| We then just create a map in Python linking the steam id to the player ID. | |||||
| ```python | |||||
| name_map = {} | |||||
| with open("friendsMap.map") as fp: | |||||
| for line in fp: | |||||
| name_map[line.split()[0]] = line.split()[1] | |||||
| ``` | |||||
| ```python | |||||
| name_map | |||||
| {'76561198188400721': 'jrtechs', | |||||
| '76561198049526995': 'Noosh', | |||||
| ... | |||||
| '76561198065642391': 'Therefore', | |||||
| '76561198121369685': 'DataFrogman'} | |||||
| ``` | |||||
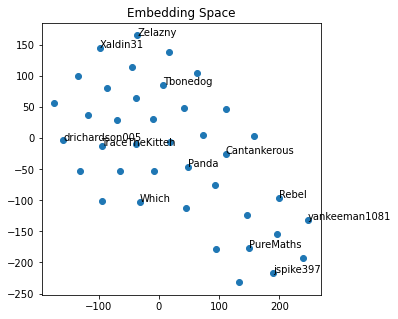
| Using the output from the TSNE dimensionality reduction, we can view all the nodes on a single plot. To make the graph look more delightful, we only label a fraction of the nodes. | |||||
| ```python | |||||
| import matplotlib.pyplot as plt | |||||
| import random | |||||
| def plot_with_matplotlib(x_vals, y_vals, labels, num_to_label): | |||||
| plt.figure(figsize=(5, 5)) | |||||
| plt.scatter(x_vals, y_vals) | |||||
| plt.title("Embedding Space") | |||||
| indices = list(range(len(labels))) | |||||
| selected_indices = random.sample(indices, num_to_label) | |||||
| for i in selected_indices: | |||||
| plt.annotate(name_map[labels[i]], (x_vals[i], y_vals[i])) | |||||
| plt.savefig('ex.png') | |||||
| plot_with_matplotlib(x_vals, y_vals, labels, 12) | |||||
| ``` | |||||
|  | |||||
| This graph may not look exciting, but I assure you that it is. | |||||
| Just eyeballing it, I can notice that my high school friends and college friends are in different regions of the graph. | |||||
| Moving forward with this, I plan on incorporating game data. | |||||
| With the graph data vectorized in this fashion, it becomes possible to start employing classification and link prediction algorithms. | |||||
| Some examples for the steam network could be a friend or game recommendation system, or even a community detection algorithm. | |||||