5 changed files with 146 additions and 0 deletions
Split View
Diff Options
-
+146 -0blogContent/posts/open-source/jupyter-will-change-your-life.md
-
BINblogContent/posts/open-source/media/jupyter/darkTheme.png
-
BINblogContent/posts/open-source/media/jupyter/light.png
-
BINblogContent/posts/open-source/media/jupyter/localForward.png
-
BINblogContent/posts/open-source/media/jupyter/network.jpeg
+ 146
- 0
blogContent/posts/open-source/jupyter-will-change-your-life.md
View File
| @ -0,0 +1,146 @@ | |||
| It is not uncommon for me to get exuberantly excited over a open source | |||
| project that I stumble upon, however, Jupyter Lab has taken the | |||
| cake this month. The Jypyter project is an open-source community | |||
| that extended IPython notebook project to the web browser and added | |||
| support for multiple languages. | |||
| # Why Notebooks? | |||
| As a researcher and educator I love notebooks because they enable | |||
| you to easily share your code with others. Notebooks are much more | |||
| interactive than simply sharing source code because you can | |||
| mix text(markdown), code, and outputs from code execution. For classes and | |||
| when working, this makes it very easy to generate quick reports. | |||
| You can simply write a document that auto generates the graphs and figures | |||
| you want to talk about in your document. | |||
| Last week I worked on a computer vision assignment that required me to | |||
| use Open CV to manipulate images using filters, convolutions, etc. | |||
| The entirety of the assignment required me to produce roughly 30 images. | |||
| A majority of the class wrote python scripts and threw each image they | |||
| generated into a massive word document and typed up their | |||
| analysis and submitted their assignment as a PDF along side a bunch of | |||
| python scripts. There is nothing wrong with doing that; however, what | |||
| happens if at the end of the assignment you realized that you were | |||
| generating Gaussian filters incorrectly? If you wrote everything in | |||
| a Jupyter notebook you would just have to fix the dubious code and | |||
| re-run the notebook and it would produce your report in its entirety. | |||
| But, if you had your scripts as separate files you would have to fix your | |||
| code and then go through and generate a dozen new images that required | |||
| Gaussian filters and place them in your document. | |||
| The ability to accurately reproduce your report is pinnacle to making | |||
| research more verifiable and reproducible. This is something that the | |||
| R and open-science communities heavily focus on. Directly mixing your | |||
| code and analysis with your report is very useful. Also, consider if the | |||
| data that you are working with changes half way through writing your | |||
| research report. With a notebook, you would just have to re-run the | |||
| notebook where if you had the report as a separate word or Latex file, | |||
| you now run the risk of misreporting your results. | |||
| # Jupyter Notebook | |||
| # Jupyter Lab | |||
| # Running and Installing | |||
| # Running for remote use | |||
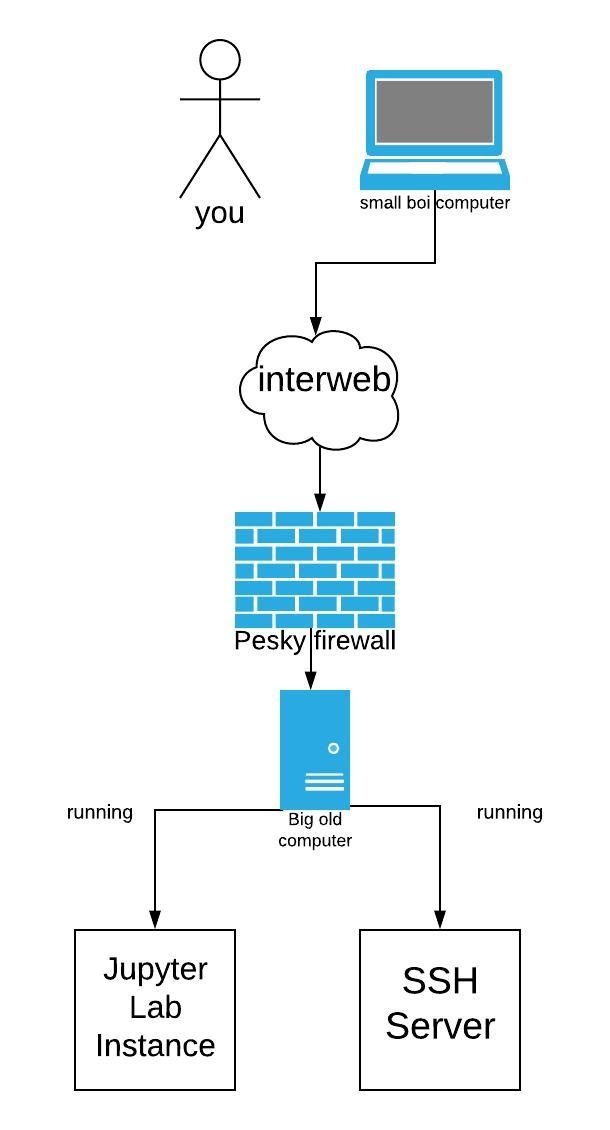
| Imagine that you are running an old computer and you simply want your | |||
| code to run on a remote computer that has a beefie GPU for ML. | |||
| With Jupyter Lab or Notebook you can do that, but, it takes a little | |||
| trickery. The easiest solution that I found involves using a reverse | |||
| SSH proxy. | |||
|  | |||
| The first thing that you want to do is set up a password so that you | |||
| can connect to the jupyter lab instance using a password rather than using | |||
| a authentication key which gets hidden in the terminal. | |||
| ```bash | |||
| jupyter notebook password | |||
| ``` | |||
| ** note ** the password that you set is configured in the same config used by both jupyter lab and jupyter notebook. | |||
| The next thing you should do is run the jupyter lab instance on the port that you want it to listen to. | |||
| ```bash | |||
| jupyter lab --no-browser --port=6000 | |||
| ``` | |||
| The "--no-browser" will prevent jupyter from opening in your default web browser. | |||
| The next step is to do a local SSH port forward on your machine | |||
| so you can access the jupyter instance on the remote server. | |||
| The benefit of doing this is that you can get behind firewalls and that | |||
| all your traffic is encrypted. | |||
|  | |||
| The image above comes from my presentation on "[Everything SSH](https://jrtechs.net/open-source/teaching-ssh-through-a-ctf)". | |||
| The essence of the command bellow is that you will forward all | |||
| connections on your machines to port 6000 to a remote's servers connection to localhost:6000. | |||
| ```bash | |||
| ssh -L 6000:localhost:6000 user@some-remote-host.rit.edu | |||
| ``` | |||
| After you run that command you can access the jupyter lab instance | |||
| by opening your favorite web client and going to localhost:6000. | |||
| Typing that command every time is tedious so I recommend that you | |||
| allias it in your shells config file. | |||
| ```bash | |||
| alias jj="ssh -L 6000:localhost:6000 user@some-remote-host.rit.edu" | |||
| ``` | |||
| Now all you have to type in your command prompt is jj to connect to | |||
| your remote jupyter server. Neat. | |||
| But, what if your roommate trips and your server gets restarted? Well, | |||
| you can write a systemd script to automatically start your jupyter | |||
| server when the computer boots. This is what my system d script looks like. | |||
| ```bash | |||
| # location /lib/systemd/system | |||
| # | |||
| # After file creation run: systemctl daemon-reload | |||
| # enable service on start up: systemctl enable jupyter-lab | |||
| # start the service: systemctl start jupyter-lab | |||
| [Unit] | |||
| Description=Script to start jupyter lab | |||
| Documentation=https://jrtechs.net | |||
| After=network.target | |||
| [Service] | |||
| Type=simple | |||
| User=jeff | |||
| WorkingDirectory=/home/jeff/Documents/school/csci-431/ | |||
| ExecStart=/usr/local/bin/jupyter lab --no-browser --port=6969 | |||
| Restart=on-failure | |||
| [Install] | |||
| WantedBy=multi-user.target | |||
| ``` | |||
| You want to set the working directory to be the location where your jupyter notebooks are stored. | |||
| You also want to make sure that you specify the absolute path to the jupyter binary in the execstart parameter. You can find that using the which command: | |||
| ```bash | |||
| which jupyter | |||
| ``` | |||