13 changed files with 1089 additions and 8 deletions
Split View
Diff Options
-
+13 -1blog/renderBlogPost.js
-
BINblogContent/headerImages/gitHubGraphs.png
-
+484 -0blogContent/posts/data-science/html/lets-build-a-genetic-algorithm.html
-
+515 -0blogContent/posts/data-science/lets-build-a-genetic-algorithm.md
-
+1 -0blogContent/posts/data-science/media/GA/GAFlowChart.svg
-
+72 -0blogContent/posts/projects/github-graphs-project.md
-
BINblogContent/posts/projects/media/github/RepositoriesView.png
-
BINblogContent/posts/projects/media/github/fossRITOrg.png
-
BINblogContent/posts/projects/media/github/jrtechsGraph.png
-
BINblogContent/posts/projects/media/github/ritlugOrg.png
-
+1 -6templates/blog/blogMain.html
-
+1 -1templates/blog/paginationBar.html
-
+2 -0templates/blog/sideBar.html
+ 13
- 1
blog/renderBlogPost.js
View File
BIN
blogContent/headerImages/gitHubGraphs.png
View File
+ 484
- 0
blogContent/posts/data-science/html/lets-build-a-genetic-algorithm.html
View File
| @ -0,0 +1,484 @@ | |||
| <script type="text/javascript" src="https://www.gstatic.com/charts/loader.js"></script> | |||
| <script | |||
| src="https://code.jquery.com/jquery-3.3.1.slim.min.js" | |||
| integrity="sha256-3edrmyuQ0w65f8gfBsqowzjJe2iM6n0nKciPUp8y+7E=" | |||
| crossorigin="anonymous"> | |||
| </script> | |||
| <script> | |||
| class Gene | |||
| { | |||
| /** | |||
| * Constructs a new Gene to store in a chromosome. | |||
| * @param min minimum value that this gene can store | |||
| * @param max value this gene can possibly be | |||
| * @param value normalized value | |||
| */ | |||
| constructor(min, max, value) | |||
| { | |||
| this.min = min; | |||
| this.max = max; | |||
| this.value = value; | |||
| } | |||
| /** | |||
| * De-normalizes the value of the gene | |||
| * @returns {*} | |||
| */ | |||
| getRealValue() | |||
| { | |||
| return (this.max - this.min) * this.value + this.min; | |||
| } | |||
| getValue() | |||
| { | |||
| return this.value; | |||
| } | |||
| setValue(val) | |||
| { | |||
| this.value = val; | |||
| } | |||
| makeClone() | |||
| { | |||
| return new Gene(this.min, this.max, this.value); | |||
| } | |||
| makeRandomGene() | |||
| { | |||
| return new Gene(this.min, this.max, Math.random()); | |||
| } | |||
| } | |||
| class Chromosome | |||
| { | |||
| /** | |||
| * Constructs a chromosome by making a copy of | |||
| * a list of genes. | |||
| * @param geneArray | |||
| */ | |||
| constructor(geneArray) | |||
| { | |||
| this.genes = []; | |||
| for(let i = 0; i < geneArray.length; i++) | |||
| { | |||
| this.genes.push(geneArray[i].makeClone()); | |||
| } | |||
| } | |||
| getGenes() | |||
| { | |||
| return this.genes; | |||
| } | |||
| /** | |||
| * Mutates a random gene. | |||
| */ | |||
| mutate() | |||
| { | |||
| this.genes[Math.round(Math.random() * (this.genes.length-1))].setValue(Math.random()); | |||
| } | |||
| /** | |||
| * Creates a totally new chromosome with same | |||
| * genetic structure as this chromosome but different | |||
| * values. | |||
| * @returns {Chromosome} | |||
| */ | |||
| createRandomChromosome() | |||
| { | |||
| let geneAr = []; | |||
| for(let i = 0; i < this.genes.length; i++) | |||
| { | |||
| geneAr.push(this.genes[i].makeRandomGene()); | |||
| } | |||
| return new Chromosome(geneAr); | |||
| } | |||
| } | |||
| /** | |||
| * Mates two chromosomes using the blending method | |||
| * and returns a list of 2 offspring. | |||
| * @param father | |||
| * @param mother | |||
| * @returns {Chromosome[]} | |||
| */ | |||
| const breed = function(father, mother) | |||
| { | |||
| let son = new Chromosome(father.getGenes()); | |||
| let daughter = new Chromosome(mother.getGenes()); | |||
| for(let i = 0;i < son.getGenes().length; i++) | |||
| { | |||
| let blendCoef = Math.random(); | |||
| blendGene(son.getGenes()[i], daughter.getGenes()[i], blendCoef); | |||
| } | |||
| return [son, daughter]; | |||
| }; | |||
| /** | |||
| * Blends two genes together based on a random blend | |||
| * coefficient. | |||
| **/ | |||
| const blendGene = function(gene1, gene2, blendCoef) | |||
| { | |||
| let value1 = (blendCoef * gene1.getValue()) + | |||
| (gene2.getValue() * (1- blendCoef)); | |||
| let value2 = ((1-blendCoef) * gene1.getValue()) + | |||
| (gene2.getValue() * blendCoef); | |||
| gene1.setValue(value1); | |||
| gene2.setValue(value2); | |||
| }; | |||
| /** | |||
| * Helper function to sort an array | |||
| * | |||
| * @param prop name of JSON property to sort by | |||
| * @returns {function(*, *): number} | |||
| */ | |||
| function predicateBy(prop) | |||
| { | |||
| return function(a,b) | |||
| { | |||
| var result; | |||
| if(a[prop] > b[prop]) | |||
| { | |||
| result = 1; | |||
| } | |||
| else if(a[prop] < b[prop]) | |||
| { | |||
| result = -1; | |||
| } | |||
| return result; | |||
| } | |||
| } | |||
| /** | |||
| * Function which computes the fitness of everyone in the | |||
| * population and returns the most fit survivors. Method | |||
| * known as elitism. | |||
| * | |||
| * @param population | |||
| * @param keepNumber | |||
| * @param fitnessFunction | |||
| * @returns {{average: number, | |||
| * survivors: Array, bestFit: Chromosome }} | |||
| */ | |||
| const naturalSelection = function(population, keepNumber, fitnessFunction) | |||
| { | |||
| let fitnessArray = []; | |||
| let total = 0; | |||
| for(let i = 0; i < population.length; i++) | |||
| { | |||
| const fitness = fitnessFunction(population[i]); | |||
| fitnessArray.push({fit:fitness, chrom: population[i]}); | |||
| total+= fitness; | |||
| } | |||
| fitnessArray.sort(predicateBy("fit")); | |||
| let survivors = []; | |||
| let bestFitness = fitnessArray[0].fit; | |||
| let bestChromosome = fitnessArray[0].chrom; | |||
| for(let i = 0; i < keepNumber; i++) | |||
| { | |||
| survivors.push(fitnessArray[i].chrom); | |||
| } | |||
| return {average: total/population.length, survivors: survivors, bestFit: bestFitness, bestChrom: bestChromosome}; | |||
| }; | |||
| /** | |||
| * Randomly everyone in the population | |||
| * | |||
| * @param population | |||
| * @param desiredPopulationSize | |||
| */ | |||
| const matePopulation = function(population, desiredPopulationSize) | |||
| { | |||
| const originalLength = population.length; | |||
| while(population.length < desiredPopulationSize) | |||
| { | |||
| let index1 = Math.round(Math.random() * (originalLength-1)); | |||
| let index2 = Math.round(Math.random() * (originalLength-1)); | |||
| if(index1 !== index2) | |||
| { | |||
| const babies = breed(population[index1], population[index2]); | |||
| population.push(babies[0]); | |||
| population.push(babies[1]); | |||
| } | |||
| } | |||
| }; | |||
| /** | |||
| * Randomly mutates the population | |||
| **/ | |||
| const mutatePopulation = function(population, mutatePercentage) | |||
| { | |||
| if(population.length >= 2) | |||
| { | |||
| let mutations = mutatePercentage * | |||
| population.length * | |||
| population[0].getGenes().length; | |||
| for(let i = 0; i < mutations; i++) | |||
| { | |||
| population[i].mutate(); | |||
| } | |||
| } | |||
| else | |||
| { | |||
| console.log("Error, population too small to mutate"); | |||
| } | |||
| }; | |||
| /** | |||
| * Introduces x random chromosomes to the population. | |||
| * @param population | |||
| * @param immigrationSize | |||
| */ | |||
| const newBlood = function(population, immigrationSize) | |||
| { | |||
| for(let i = 0; i < immigrationSize; i++) | |||
| { | |||
| let geneticChromosome = population[0]; | |||
| population.push(geneticChromosome.createRandomChromosome()); | |||
| } | |||
| }; | |||
| let costx = Math.random() * 10; | |||
| let costy = Math.random() * 10; | |||
| /** Defines the cost as the "distance" to a 2-d point. | |||
| * @param chromosome | |||
| * @returns {number} | |||
| */ | |||
| const basicCostFunction = function(chromosome) | |||
| { | |||
| return Math.abs(chromosome.getGenes()[0].getRealValue() - costx) + | |||
| Math.abs(chromosome.getGenes()[1].getRealValue() - costy); | |||
| }; | |||
| /** | |||
| * Creates a totally random population based on a desired size | |||
| * and a prototypical chromosome. | |||
| * | |||
| * @param geneticChromosome | |||
| * @param populationSize | |||
| * @returns {Array} | |||
| */ | |||
| const createRandomPopulation = function(geneticChromosome, populationSize) | |||
| { | |||
| let population = []; | |||
| for(let i = 0; i < populationSize; i++) | |||
| { | |||
| population.push(geneticChromosome.createRandomChromosome()); | |||
| } | |||
| return population; | |||
| }; | |||
| /** | |||
| * Runs the genetic algorithm by going through the processes of | |||
| * natural selection, mutation, mating, and immigrations. This | |||
| * process will continue until an adequately performing chromosome | |||
| * is found or a generation threshold is passed. | |||
| * | |||
| * @param geneticChromosome Prototypical chromosome: used so algo knows | |||
| * what the dna of the population looks like. | |||
| * @param costFunction Function which defines how bad a Chromosome is | |||
| * @param populationSize Desired population size for population | |||
| * @param maxGenerations Cut off level for number of generations to run | |||
| * @param desiredCost Sufficient cost to terminate program at | |||
| * @param mutationRate Number between [0,1] representing proportion of genes | |||
| * to mutate each generation | |||
| * @param keepNumber Number of Organisms which survive each generation | |||
| * @param newBloodNumber Number of random immigrants to introduce into | |||
| * the population each generation. | |||
| * @returns {*} | |||
| */ | |||
| const runGeneticOptimization = function(geneticChromosome, costFunction, | |||
| populationSize, maxGenerations, | |||
| desiredCost, mutationRate, keepNumber, | |||
| newBloodNumber) | |||
| { | |||
| let population = createRandomPopulation(geneticChromosome, populationSize); | |||
| let generation = 0; | |||
| let bestCost = Number.MAX_VALUE; | |||
| let bestChromosome = geneticChromosome; | |||
| do | |||
| { | |||
| matePopulation(population, populationSize); | |||
| newBlood(population, newBloodNumber); | |||
| mutatePopulation(population, mutationRate); | |||
| let generationResult = naturalSelection(population, keepNumber, costFunction); | |||
| if(bestCost > generationResult.bestFit) | |||
| { | |||
| bestChromosome = generationResult.bestChrom; | |||
| bestCost = generationResult.bestFit; | |||
| } | |||
| population = generationResult.survivors; | |||
| generation++; | |||
| console.log("Generation " + generation + " Best Cost: " + bestCost); | |||
| }while(generation < maxGenerations && bestCost > desiredCost); | |||
| return bestChromosome; | |||
| }; | |||
| /** | |||
| * Ugly globals used to keep track of population state for the graph. | |||
| */ | |||
| let genericChromosomeG, costFunctionG, | |||
| populationSizeG, maxGenerationsG, | |||
| desiredCostG, mutationRateG, keepNumberG, | |||
| newBloodNumberG, populationG, generationG, | |||
| bestCostG = Number.MAX_VALUE, bestChromosomeG = genericChromosomeG; | |||
| const runGeneticOptimizationForGraph = function() | |||
| { | |||
| let generationResult = naturalSelection(populationG, keepNumberG, costFunctionG); | |||
| stats.push([generationG, generationResult.bestFit, generationResult.average]); | |||
| if(bestCostG > generationResult.bestFit) | |||
| { | |||
| bestChromosomeG = generationResult.bestChrom; | |||
| bestCostG = generationResult.bestFit; | |||
| } | |||
| populationG = generationResult.survivors; | |||
| generationG++; | |||
| console.log("Generation " + generationG + " Best Cost: " + bestCostG); | |||
| console.log(generationResult); | |||
| matePopulation(populationG, populationSizeG); | |||
| newBlood(populationG, newBloodNumberG); | |||
| mutatePopulation(populationG, mutationRateG); | |||
| createGraph(); | |||
| }; | |||
| let stats = []; | |||
| const createGraph = function() | |||
| { | |||
| var dataPoints = []; | |||
| console.log(dataPoints); | |||
| var data = new google.visualization.DataTable(); | |||
| data.addColumn('number', 'Gene 1'); | |||
| data.addColumn('number', 'Gene 2'); | |||
| for(let i = 0; i < populationG.length; i++) | |||
| { | |||
| data.addRow([populationG[i].getGenes()[0].getRealValue(), | |||
| populationG[i].getGenes()[1].getRealValue()]); | |||
| } | |||
| var options = { | |||
| title: 'Genetic Evolution On Two Genes Generation: ' + generationG, | |||
| hAxis: {title: 'Gene 1', minValue: 0, maxValue: 10}, | |||
| vAxis: {title: 'Gene 2', minValue: 0, maxValue: 10}, | |||
| }; | |||
| var chart = new google.visualization.ScatterChart(document.getElementById('chart_div')); | |||
| chart.draw(data, options); | |||
| //line chart stuff | |||
| var line_data = new google.visualization.DataTable(); | |||
| line_data.addColumn('number', 'Generation'); | |||
| line_data.addColumn('number', 'Best'); | |||
| line_data.addColumn('number', 'Average'); | |||
| line_data.addRows(stats); | |||
| console.log(stats); | |||
| var lineChartOptions = { | |||
| hAxis: { | |||
| title: 'Generation' | |||
| }, | |||
| vAxis: { | |||
| title: 'Cost' | |||
| }, | |||
| colors: ['#AB0D06', '#007329'] | |||
| }; | |||
| var chart = new google.visualization.LineChart(document.getElementById('line_chart')); | |||
| chart.draw(line_data, lineChartOptions); | |||
| }; | |||
| let gene1 = new Gene(1,10,10); | |||
| let gene2 = new Gene(1,10,0.4); | |||
| let geneList = [gene1, gene2]; | |||
| let exampleOrganism = new Chromosome(geneList); | |||
| genericChromosomeG = exampleOrganism; | |||
| costFunctionG = basicCostFunction; | |||
| populationSizeG = 100; | |||
| maxGenerationsG = 30; | |||
| desiredCostG = 0.00001; | |||
| mutationRateG = 0.3; | |||
| keepNumberG = 30; | |||
| newBloodNumberG = 10; | |||
| generationG = 0; | |||
| function verifyForm() | |||
| { | |||
| if(Number($("#populationSize").val()) <= 1) | |||
| { | |||
| alert("Population size must be greater than one."); | |||
| return false; | |||
| } | |||
| if(Number($("#mutationRate").val()) > 1 || | |||
| Number($("#mutationRate").val()) < 0) | |||
| { | |||
| alert("Mutation rate must be between zero and one."); | |||
| return false; | |||
| } | |||
| if(Number($("#survivalSize").val()) < 0) | |||
| { | |||
| alert("Survival size can't be less than one."); | |||
| return false; | |||
| } | |||
| if(Number($("#newBlood").val()) < 0) | |||
| { | |||
| alert("New organisms can't be a negative number."); | |||
| return false; | |||
| } | |||
| return true; | |||
| } | |||
| function resetPopulation() | |||
| { | |||
| if(verifyForm()) | |||
| { | |||
| stats = []; | |||
| autoRunning = false; | |||
| $("#runAutoOptimizer").val("Auto Run"); | |||
| populationSizeG = $("#populationSize").val(); | |||
| mutationRateG = $("#mutationRate").val(); | |||
| keepNumberG = $("#survivalSize").val(); | |||
| newBloodNumberG = $("#newBlood").val(); | |||
| generationG = 0; | |||
| populationG = createRandomPopulation(genericChromosomeG, populationSizeG); | |||
| createGraph(); | |||
| } | |||
| } | |||
| populationG = createRandomPopulation(genericChromosomeG, populationSizeG); | |||
| window.onload = function (){ | |||
| google.charts.load('current', {packages: ['corechart', 'line']}); | |||
| google.charts.load('current', {'packages':['corechart']}).then(function() | |||
| { | |||
| createGraph(); | |||
| }) | |||
| }; | |||
| let autoRunning = false; | |||
| function runAutoOptimizer() | |||
| { | |||
| if(autoRunning === true) | |||
| { | |||
| runGeneticOptimizationForGraph(); | |||
| setTimeout(runAutoOptimizer, 1000); | |||
| } | |||
| } | |||
| function startStopAutoRun() | |||
| { | |||
| autoRunning = !autoRunning; | |||
| if(autoRunning) | |||
| { | |||
| $("#runAutoOptimizer").val("Stop Auto Run"); | |||
| } | |||
| else | |||
| { | |||
| $("#runAutoOptimizer").val("Resume Auto Run"); | |||
| } | |||
| runAutoOptimizer(); | |||
| } | |||
| </script> | |||
| <div id="chart_div"></div> | |||
| <div id="line_chart"></div> | |||
| <input class='btn btn-primary' id="runOptimizer" onclick='runGeneticOptimizationForGraph()' type="button" value="Next Generation"> | |||
| <input class='btn btn-primary' id="runAutoOptimizer" onclick='startStopAutoRun()' type="button" value="Auto Run"> | |||
| <br> | |||
| <br> | |||
| <div class="card"> | |||
| <div class="card-header"> | |||
| <h2>Population Variables</h2> | |||
| </div> | |||
| <form class="card-body"> | |||
| <div class="row p-2"> | |||
| <div class="col"> | |||
| <label for="populationSize">Population Size</label> | |||
| <input type="text" class="form-control" value="100" id="populationSize" placeholder="Population Size" required> | |||
| </div> | |||
| <div class="col"> | |||
| <label for="populationSize">Survival Size</label> | |||
| <input type="text" class="form-control" value="20" id="survivalSize" placeholder="Survival Size" required> | |||
| </div> | |||
| </div> | |||
| <div class="row p-2"> | |||
| <div class="col"> | |||
| <label for="populationSize">Mutation Rate</label> | |||
| <input type="text" class="form-control" value="0.03" id="mutationRate" placeholder="Mutation Rate" required> | |||
| </div> | |||
| <div class="col"> | |||
| <label for="populationSize">New Organisms Per Generation</label> | |||
| <input type="text" class="form-control" value="5" id="newBlood" placeholder="New Organisms" required> | |||
| </div> | |||
| </div> | |||
| <br> | |||
| <input class='btn btn-primary' id="reset" onclick='resetPopulation()' type="button" value="Reset Population"> | |||
| </form> | |||
| </div> | |||
+ 515
- 0
blogContent/posts/data-science/lets-build-a-genetic-algorithm.md
View File
| @ -0,0 +1,515 @@ | |||
| # Live Simulation | |||
| <customHTML /> | |||
| # Background and Theory | |||
| Since you stumbled upon this article, you might be wondering what the heck genetic algorithms are. | |||
| To put it simply: genetic algorithms employ the same tactics used in natural selection to find an optimal solution to an optimization problem. | |||
| Genetic algorithms are often used in high dimensional problems where the optimal solutions are not apparent. | |||
| Genetic algorithms are commonly used to tune the [hyper-parameters](https://en.wikipedia.org/wiki/Hyperparameter) of a program. | |||
| However, this algorithm can be used in any scenario where you have a function which defines how well a solution is. | |||
| Many people have used genetic algorithms in video games to auto learn the weaknesses of players. | |||
| The beautiful part about Genetic Algorithms are their simplicity; you need absolutely no knowledge of linear algebra or calculus. | |||
| To implement a genetic algorithm from scratch you only need **very basic** algebra and a general grasp of evolution. | |||
| # Genetic Algorithm | |||
| All genetic algorithms typically have a single cycle where you continuously mutate, breed, and select the most optimal solutions. | |||
| I will dive into each section of this algorithm using simple JavaScript code snippets. | |||
| The algorithm which I present is very generic and modular so it should be easy to port into other programming languages and applications. | |||
|  | |||
| ## Population Creation | |||
| The very first thing we need to do is specify a data-structure for storing our genetic information. | |||
| In biology, chromosomes are composed of sequences of genes. | |||
| Many people run genetic algorithms on binary arrays since they more closely represent DNA. | |||
| However, as computer scientists, it is often easier to model problems using continuous numbers. | |||
| In this approach, every gene will be a single floating point number ranging between zero and one. | |||
| Every type of gene will have a max and min value which represents the absolute extremes of that gene. | |||
| This works well for optimization because it allows us to easily limit our search space. | |||
| For example, we can specify that "height" gene can only vary between 0 and 90. | |||
| To get the actual value of the gene from its \[0-1] value we simple de-normalize it. | |||
| $$ | |||
| g_{real value} = (g_{high}- g_{low})g_{norm} + g_{low} | |||
| $$ | |||
| ```javascript | |||
| class Gene | |||
| { | |||
| /** | |||
| * Constructs a new Gene to store in a chromosome. | |||
| * @param min minimum value that this gene can store | |||
| * @param max value this gene can possibly be | |||
| * @param value normalized value | |||
| */ | |||
| constructor(min, max, value) | |||
| { | |||
| this.min = min; | |||
| this.max = max; | |||
| this.value = value; | |||
| } | |||
| /** | |||
| * De-normalizes the value of the gene | |||
| * @returns {*} | |||
| */ | |||
| getRealValue() | |||
| { | |||
| return (this.max - this.min) * this.value + this.min; | |||
| } | |||
| getValue() | |||
| { | |||
| return this.value; | |||
| } | |||
| setValue(val) | |||
| { | |||
| this.value = val; | |||
| } | |||
| makeClone() | |||
| { | |||
| return new Gene(this.min, this.max, this.value); | |||
| } | |||
| makeRandomGene() | |||
| { | |||
| return new Gene(this.min, this.max, Math.random()); | |||
| } | |||
| } | |||
| ``` | |||
| Now that we have genes, we can create chromosomes. | |||
| Chromosomes are simply collections of genes. | |||
| Whatever language you make this in, make sure that when you create a new chromosome it | |||
| is has a [deep copy](https://en.wikipedia.org/wiki/Object_copying) of the original genetic information rather than a shallow copy. | |||
| A shallow copy is when you simple copy the object pointer where a deep copy is actually creating a new object. | |||
| If you fail to do a deep copy, you will have weird issues where multiple chromosomes will share the same DNA. | |||
| In this class I added helper functions to clone the chromosome as a random copy. | |||
| You can only create a new chromosome by cloning because I wanted to keep the program generic and make no assumptions about the domain. | |||
| Since you only provide the min/max information for the genes once, cloning an existing chromosome is the easiest way of | |||
| ensuring that all corresponding chromosomes contain genes with identical extrema. | |||
| ```javascript | |||
| class Chromosome | |||
| { | |||
| /** | |||
| * Constructs a chromosome by making a copy of | |||
| * a list of genes. | |||
| * @param geneArray | |||
| */ | |||
| constructor(geneArray) | |||
| { | |||
| this.genes = []; | |||
| for(let i = 0; i < geneArray.length; i++) | |||
| { | |||
| this.genes.push(geneArray[i].makeClone()); | |||
| } | |||
| } | |||
| getGenes() | |||
| { | |||
| return this.genes; | |||
| } | |||
| /** | |||
| * Mutates a random gene. | |||
| */ | |||
| mutate() | |||
| { | |||
| this.genes[Math.round(Math.random() * (this.genes.length-1))].setValue(Math.random()); | |||
| } | |||
| /** | |||
| * Creates a totally new chromosome with same | |||
| * genetic structure as this chromosome but different | |||
| * values. | |||
| * @returns {Chromosome} | |||
| */ | |||
| createRandomChromosome() | |||
| { | |||
| let geneAr = []; | |||
| for(let i = 0; i < this.genes.length; i++) | |||
| { | |||
| geneAr.push(this.genes[i].makeRandomGene()); | |||
| } | |||
| return new Chromosome(geneAr); | |||
| } | |||
| } | |||
| ``` | |||
| Creating a random population is pretty straight forward if implemented a method to create a random clone of a chromosome. | |||
| ```javascript | |||
| /** | |||
| * Creates a totally random population based on a desired size | |||
| * and a prototypical chromosome. | |||
| * | |||
| * @param geneticChromosome | |||
| * @param populationSize | |||
| * @returns {Array} | |||
| */ | |||
| const createRandomPopulation = function(geneticChromosome, populationSize) | |||
| { | |||
| let population = []; | |||
| for(let i = 0; i < populationSize; i++) | |||
| { | |||
| population.push(geneticChromosome.createRandomChromosome()); | |||
| } | |||
| return population; | |||
| }; | |||
| ``` | |||
| This is where nearly all the domain information is introduced. | |||
| After you define what types of genes are found on each chromosome, you can create an entire population. | |||
| In this example all genes contain values ranging between one and ten. | |||
| ```javascript | |||
| let gene1 = new Gene(1,10,10); | |||
| let gene2 = new Gene(1,10,0.4); | |||
| let geneList = [gene1, gene2]; | |||
| let exampleOrganism = new Chromosome(geneList); | |||
| let population = createRandomPopulation(genericChromosome, 100); | |||
| ``` | |||
| ## Evaluate Fitness | |||
| Like all optimization problems, you need a way to evaluate the performance of a particular solution. | |||
| The cost function takes in a chromosome and evaluates how close it got to the ideal solution. | |||
| This particular example it is just computing the [Manhattan Distance](https://en.wiktionary.org/wiki/Manhattan_distance) to a random 2D point. | |||
| I chose two dimensions because it is easy to graph, however, real applications may have dozens of genes on each chromosome. | |||
| ```javascript | |||
| let costx = Math.random() * 10; | |||
| let costy = Math.random() * 10; | |||
| /** Defines the cost as the "distance" to a 2-d point. | |||
| * @param chromosome | |||
| * @returns {number} | |||
| */ | |||
| const basicCostFunction = function(chromosome) | |||
| { | |||
| return Math.abs(chromosome.getGenes()[0].getRealValue() - costx) + | |||
| Math.abs(chromosome.getGenes()[1].getRealValue() - costy); | |||
| }; | |||
| ``` | |||
| ## Selection | |||
| Selecting the best performing chromosomes is straightforward after you have a function for evaluating the performance. | |||
| This code snippet also computes the average and best chromosome of the population to make it easier to graph and define | |||
| the stopping point for the algorithm's main loop. | |||
| ```javascript | |||
| /** | |||
| * Function which computes the fitness of everyone in the | |||
| * population and returns the most fit survivors. Method | |||
| * known as elitism. | |||
| * | |||
| * @param population | |||
| * @param keepNumber | |||
| * @param fitnessFunction | |||
| * @returns {{average: number, | |||
| * survivors: Array, bestFit: Chromosome }} | |||
| */ | |||
| const naturalSelection = function(population, keepNumber, fitnessFunction) | |||
| { | |||
| let fitnessArray = []; | |||
| let total = 0; | |||
| for(let i = 0; i < population.length; i++) | |||
| { | |||
| const fitness = fitnessFunction(population[i]); | |||
| fitnessArray.push({fit:fitness, chrom: population[i]}); | |||
| total+= fitness; | |||
| } | |||
| fitnessArray.sort(predicateBy("fit")); | |||
| let survivors = []; | |||
| let bestFitness = fitnessArray[0].fit; | |||
| let bestChromosome = fitnessArray[0].chrom; | |||
| for(let i = 0; i < keepNumber; i++) | |||
| { | |||
| survivors.push(fitnessArray[i].chrom); | |||
| } | |||
| return {average: total/population.length, survivors: survivors, bestFit: bestFitness, bestChrom: bestChromosome}; | |||
| }; | |||
| ``` | |||
| You might be wondering how I sorted the list of JSON objects - not a numerical array. | |||
| I used the following function as a comparator for JavaScript's built in sort function. | |||
| This comparator will compare objects based on a specific attribute that you give it. | |||
| This is a very handy function to include in all of your JavaScript projects for easy sorting. | |||
| ```javascript | |||
| /** | |||
| * Helper function to sort an array | |||
| * | |||
| * @param prop name of JSON property to sort by | |||
| * @returns {function(*, *): number} | |||
| */ | |||
| function predicateBy(prop) | |||
| { | |||
| return function(a,b) | |||
| { | |||
| var result; | |||
| if(a[prop] > b[prop]) | |||
| { | |||
| result = 1; | |||
| } | |||
| else if(a[prop] < b[prop]) | |||
| { | |||
| result = -1; | |||
| } | |||
| return result; | |||
| } | |||
| } | |||
| ``` | |||
| ## Reproduction | |||
| The process of reproduction can be broken down into Pairing and Mating. | |||
| ### Pairing | |||
| Pairing is the process of selecting mates to produce offspring. | |||
| A typical approach will separate the population into two segments of mothers and fathers. | |||
| You then randomly pick pairs of mothers and fathers to produce offspring. | |||
| It is ok if one chromosome mates more than once. | |||
| It is just important that you keep this process random. | |||
| ```javascript | |||
| /** | |||
| * Randomly everyone in the population | |||
| * | |||
| * @param population | |||
| * @param desiredPopulationSize | |||
| */ | |||
| const matePopulation = function(population, desiredPopulationSize) | |||
| { | |||
| const originalLength = population.length; | |||
| while(population.length < desiredPopulationSize) | |||
| { | |||
| let index1 = Math.round(Math.random() * (originalLength-1)); | |||
| let index2 = Math.round(Math.random() * (originalLength-1)); | |||
| if(index1 !== index2) | |||
| { | |||
| const babies = breed(population[index1], population[index2]); | |||
| population.push(babies[0]); | |||
| population.push(babies[1]); | |||
| } | |||
| } | |||
| }; | |||
| ``` | |||
| ### Mating | |||
| Mating is the actual act of forming new chromosomes/organisms based on your previously selected pairs. | |||
| From my research, there are two major forms of mating: blending, crossover. | |||
| Blending is typically the most preferred approach to mating when dealing with continuous variables. | |||
| In this approach you combine the genes of both parents based on a random factor. | |||
| $$ | |||
| c_{new} = r * c_{mother} + (1-r) * c_{father} | |||
| $$ | |||
| The second offspring simply uses (1-r) for their random factor to adjust the chromosomes. | |||
| Crossover is the simplest approach to mating. | |||
| In this process you clone the parents and then you randomly swap *n* of their genes. | |||
| This works fine in some scenarios; however, this severely lacks the genetic diversity of the genes because you now have to solely | |||
| rely on mutations for changes. | |||
| ```javascript | |||
| /** | |||
| * Mates two chromosomes using the blending method | |||
| * and returns a list of 2 offspring. | |||
| * @param father | |||
| * @param mother | |||
| * @returns {Chromosome[]} | |||
| */ | |||
| const breed = function(father, mother) | |||
| { | |||
| let son = new Chromosome(father.getGenes()); | |||
| let daughter = new Chromosome(mother.getGenes()); | |||
| for(let i = 0;i < son.getGenes().length; i++) | |||
| { | |||
| let blendCoef = Math.random(); | |||
| blendGene(son.getGenes()[i], daughter.getGenes()[i], blendCoef); | |||
| } | |||
| return [son, daughter]; | |||
| }; | |||
| /** | |||
| * Blends two genes together based on a random blend | |||
| * coefficient. | |||
| **/ | |||
| const blendGene = function(gene1, gene2, blendCoef) | |||
| { | |||
| let value1 = (blendCoef * gene1.getValue()) + | |||
| (gene2.getValue() * (1- blendCoef)); | |||
| let value2 = ((1-blendCoef) * gene1.getValue()) + | |||
| (gene2.getValue() * blendCoef); | |||
| gene1.setValue(value1); | |||
| gene2.setValue(value2); | |||
| }; | |||
| ``` | |||
| ## Mutation | |||
| Mutations are random changes to an organisms DNA. | |||
| In the scope of genetic algorithms, it helps our population converge on the correct solution. | |||
| You can either adjust genes by a factor resulting in a smaller change or, you can | |||
| change the value of the gene to be something completely random. | |||
| Since we are using the blending technique for reproduction, we already have small incremental changes. | |||
| I prefer to use mutations to randomly change the entire gene since it helps prevent the algorithm | |||
| from settling on a local minimum rather than the global minimum. | |||
| ```javascript | |||
| /** | |||
| * Randomly mutates the population | |||
| **/ | |||
| const mutatePopulation = function(population, mutatePercentage) | |||
| { | |||
| if(population.length >= 2) | |||
| { | |||
| let mutations = mutatePercentage * | |||
| population.length * | |||
| population[0].getGenes().length; | |||
| for(let i = 0; i < mutations; i++) | |||
| { | |||
| population[i].mutate(); | |||
| } | |||
| } | |||
| else | |||
| { | |||
| console.log("Error, population too small to mutate"); | |||
| } | |||
| }; | |||
| ``` | |||
| ## Immigration | |||
| Immigration or "new blood" is the process of dumping random organisms into your population at each generation. | |||
| This prevents us from getting stuck in a local minimum rather than the global minimum. | |||
| There are more advanced techniques to accomplish this same concept. | |||
| My favorite approach (not implemented here) is raising **x** populations simultaneously and every **y** generations | |||
| you take **z** organisms from each population and move them to another population. | |||
| ```javascript | |||
| /** | |||
| * Introduces x random chromosomes to the population. | |||
| * @param population | |||
| * @param immigrationSize | |||
| */ | |||
| const newBlood = function(population, immigrationSize) | |||
| { | |||
| for(let i = 0; i < immigrationSize; i++) | |||
| { | |||
| let geneticChromosome = population[0]; | |||
| population.push(geneticChromosome.createRandomChromosome()); | |||
| } | |||
| }; | |||
| ``` | |||
| ## Putting It All Together | |||
| Now that we have all the ingredients for a genetic algorithm we can piece it together in a simple loop. | |||
| ```javascript | |||
| /** | |||
| * Runs the genetic algorithm by going through the processes of | |||
| * natural selection, mutation, mating, and immigrations. This | |||
| * process will continue until an adequately performing chromosome | |||
| * is found or a generation threshold is passed. | |||
| * | |||
| * @param geneticChromosome Prototypical chromosome: used so algo knows | |||
| * what the dna of the population looks like. | |||
| * @param costFunction Function which defines how bad a Chromosome is | |||
| * @param populationSize Desired population size for population | |||
| * @param maxGenerations Cut off level for number of generations to run | |||
| * @param desiredCost Sufficient cost to terminate program at | |||
| * @param mutationRate Number between [0,1] representing proportion of genes | |||
| * to mutate each generation | |||
| * @param keepNumber Number of Organisms which survive each generation | |||
| * @param newBloodNumber Number of random immigrants to introduce into | |||
| * the population each generation. | |||
| * @returns {*} | |||
| */ | |||
| const runGeneticOptimization = function(geneticChromosome, costFunction, | |||
| populationSize, maxGenerations, | |||
| desiredCost, mutationRate, keepNumber, | |||
| newBloodNumber) | |||
| { | |||
| let population = createRandomPopulation(geneticChromosome, populationSize); | |||
| let generation = 0; | |||
| let bestCost = Number.MAX_VALUE; | |||
| let bestChromosome = geneticChromosome; | |||
| do | |||
| { | |||
| matePopulation(population, populationSize); | |||
| newBlood(population, newBloodNumber); | |||
| mutatePopulation(population, mutationRate); | |||
| let generationResult = naturalSelection(population, keepNumber, costFunction); | |||
| if(bestCost > generationResult.bestFit) | |||
| { | |||
| bestChromosome = generationResult.bestChrom; | |||
| bestCost = generationResult.bestFit; | |||
| } | |||
| population = generationResult.survivors; | |||
| generation++; | |||
| console.log("Generation " + generation + " Best Cost: " + bestCost); | |||
| }while(generation < maxGenerations && bestCost > desiredCost); | |||
| return bestChromosome; | |||
| }; | |||
| ``` | |||
| ## Running | |||
| Running the program is pretty straight forward after you have your genes and cost function defined. | |||
| You might be wondering if there is an optimal configuration of parameters to use with this algorithm. | |||
| The answer is that it varies based on the particular problem. | |||
| Problems like the one graphed by this website perform very well with a low mutation rate and a high population. | |||
| However, some higher dimensional problems won't even converge on a local answer if you set your mutation rate too low. | |||
| ```javascript | |||
| let gene1 = new Gene(1,10,10); | |||
| ... | |||
| let geneN = new Gene(1,10,0.4); | |||
| let geneList = [gene1,..., geneN]; | |||
| let exampleOrganism = new Chromosome(geneList); | |||
| costFunction = function(chromosome) | |||
| { | |||
| var d =...; | |||
| //compute cost | |||
| return d; | |||
| } | |||
| runGeneticOptimization(exampleOrganism, costFunction, 100, 50, 0.01, 0.3, 20, 10); | |||
| ``` | |||
| The complete code for the genetic algorithm and the fancy JavaScript graphs can be found in my [Random Scripts GitHub Repository](https://github.com/jrtechs/RandomScripts). | |||
| In the future I may package this into an [npm](https://www.npmjs.com/) package. | |||
+ 1
- 0
blogContent/posts/data-science/media/GA/GAFlowChart.svg
File diff suppressed because it is too large
View File
+ 72
- 0
blogContent/posts/projects/github-graphs-project.md
View File
| @ -0,0 +1,72 @@ | |||
| Shortly after working on my [Steam Friends Graph](https://jrtechs.net/projects/steam-friends-graph) | |||
| ,I had the idea of extending the project to include the GitHub network. | |||
| I used [BrickHack V](https://brickhack.io/) as the opportunity to work on this project with my friends. | |||
| Rather than simply use the code that was used in the Steam friends graph, the architecture was completely | |||
| revamped to reflect both the differences between the Steam and GitHub networks and my improved web development skills. | |||
| # Project Overview | |||
| We created an interactive website which allows you to make graphs based on the Github network. | |||
| Currently the site generates three types of graphs-- the most popular and entertaining of which is the friends graph. | |||
| The friends graph helps you visualize clusters of friends/collaborators on GitHub. | |||
| Similar to the Steam Friends Project, I hope that this project will make people more interested in learning about big data. | |||
| The visual aspect of this website makes learning about topics such as clustering and graph databases more intuitive. | |||
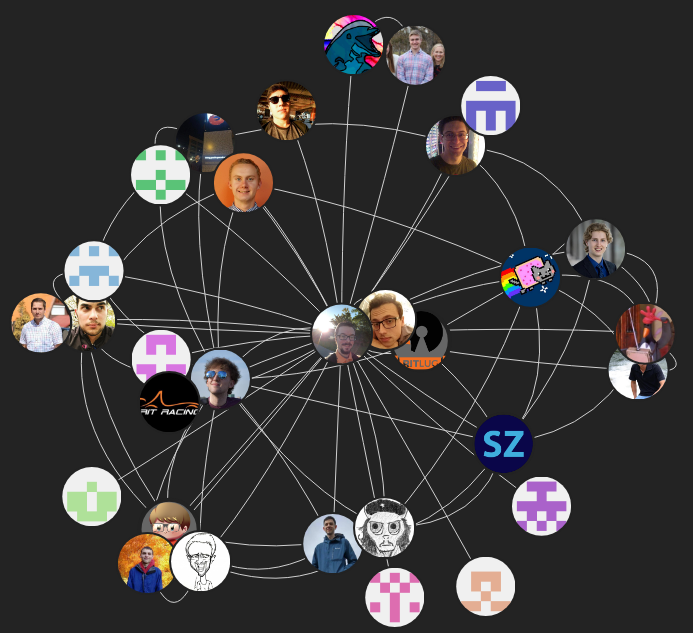
| ## Friends View | |||
|  | |||
| The friends view displays all of the people which you following and your followers. | |||
| This also connects connects everyone in the graph which are following each other. | |||
| ## Repository View | |||
|  | |||
| ## Organization View | |||
|  | |||
|  | |||
| ## Technologies Used | |||
| - [BootStrap](https://getbootstrap.com/) | |||
| - [jQuery](https://jquery.com/) | |||
| - [Vis JS](http://visjs.org/) | |||
| - [Github v3 API](https://developer.github.com/v3/) | |||
| - [Node.js](https://nodejs.org/en/) | |||
| # Changes From the Steam Graph Project | |||
| The one stark difference between the Steam network and GitHub is the amount of friends that people have. | |||
| Most developers on GitHub typically only follows around 20 people where it is not uncommon for people on Steam to have well over 100 friends. | |||
| Due to the smaller graphs, I was able to use VisJS which has nicer animations and supports custom HTML for each node. | |||
|  | |||
| Another big change to the architecture was the way in which graphs are sent to the client. | |||
| The server generated the graph and then sent the nodes and edges to the client over a web socket for the steam graph. | |||
| In this project, the client builds the graph and queries the server using ajax for the necessary information. | |||
| This gives the client a more dynamic loading progress and makes hosting the application much easier. | |||
| # Future Work | |||
| Since this project was initially created during a hackathon, there is a **lot** of work to be done. | |||
| I will outline a few ideas which I have. | |||
| - Improved Caching and Performance | |||
| - Friends of Friends -- similar to Steam's graph | |||
| - Graphs Linking Users and Repositories Based on Activity | |||
| - Code Metrics | |||
| # Contributing | |||
| If you want to contribute to this project and don't know where to start, look at the open issues on [GitHub](https://github.com/jrtechs/github-graphs). | |||
| Once you know what you want to work on, just discuss it in the issues and file a pull request. | |||
| I are very open to new contributes. | |||
| <youtube src="rz7KD_d-uQg" /> | |||